7TK4
 
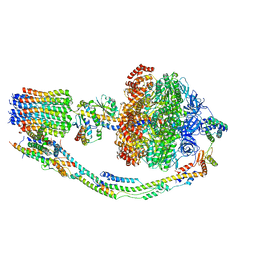 | |
7TKC
 
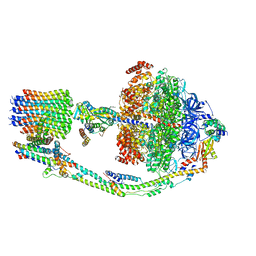 | |
1L6T
 
 | |
1L7H
 
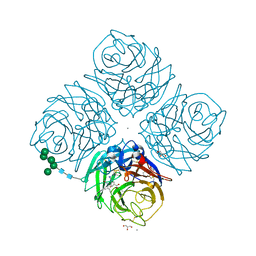 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
7UI5
 
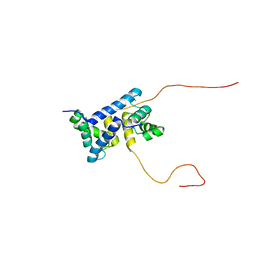 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8TLS
 
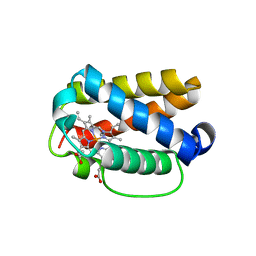 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant
To Be Published
|
|
7WXI
 
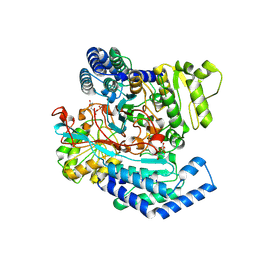 | | GPR domain of Drosophila P5CS filament with glutamate and ATPgammaS | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXF
 
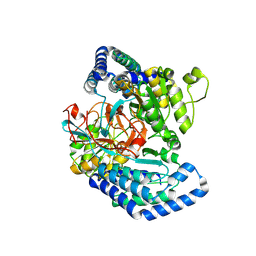 | |
7WXH
 
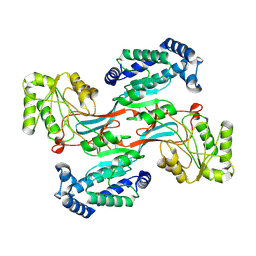 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
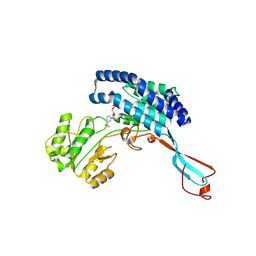 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX4
 
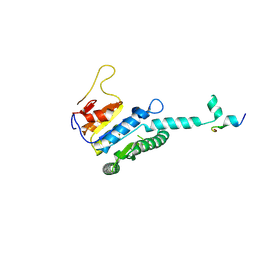 | |
7WX3
 
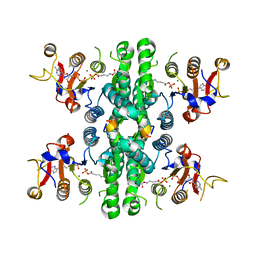 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
1I9K
 
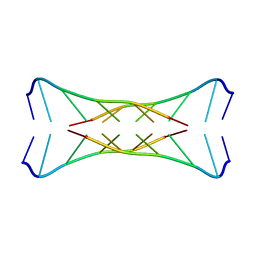 | |
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
1V11
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
7WJ4
 
 | |
1EXV
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1V1R
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, CHLORIDE ION, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
7XMV
 
 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A(AMP/ADP) filament bound with ADP, AMP and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XN3
 
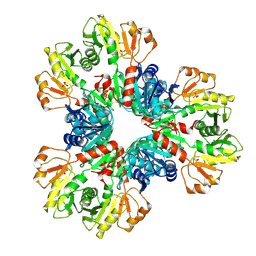 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type B filament bound with Pi | | Descriptor: | PHOSPHATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XMU
 
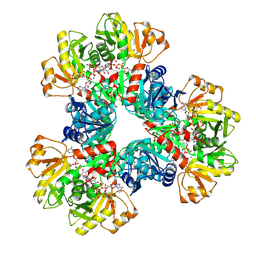 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filament bound with ADP, Pi and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
1V16
 
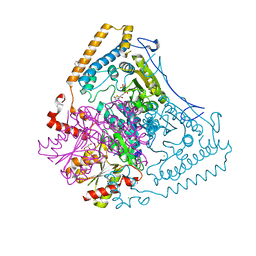 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-07 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1FVJ
 
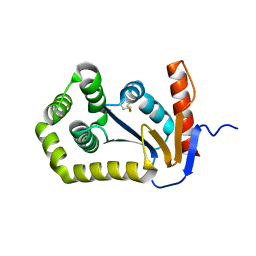 | |
1JL0
 
 | | Structure of a Human S-Adenosylmethionine Decarboxylase Self-processing Ester Intermediate and Mechanism of Putrescine Stimulation of Processing as Revealed by the H243A Mutant | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYLMETHIONINE DECARBOXYLASE PROENZYME | | Authors: | Ekstrom, J.L, Tolbert, W.D, Xiong, H, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-07-13 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a human S-adenosylmethionine decarboxylase self-processing ester intermediate and mechanism of putrescine stimulation of processing as revealed by the H243A mutant.
Biochemistry, 40, 2001
|
|
