2NRV
 
 | |
2I34
 
 | |
2I3Z
 
 | | rat DPP-IV with xanthine mimetic inhibitor #7 | | Descriptor: | 2-[(3S)-3-AMINOPIPERIDIN-1-YL]-1-(2-CYANOBENZYL)-5-METHYL-4,6-DIOXO-3,4,5,6-TETRAHYDROPYRROLO[3,4-D]IMIDAZOL-1-IUM, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Kurukulasuriya, R, Rohde, J.J, Szczepankiewicz, B.G, Basha, F, Lai, C, Winn, M, Stewart, K.D, Longenecker, K.L, Lubben, T.W, Ballaron, S.J, Sham, H.L, VonGeldern, T.W. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Xanthine mimetics as potent dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5KOX
 
 | |
2O8F
 
 | | human MutSalpha (MSH2/MSH6) bound to DNA with a single base T insert | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*CP*GP*TP*C)-3', 5'-D(*GP*AP*CP*GP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2NRR
 
 | |
2I9K
 
 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | Descriptor: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
2NRX
 
 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | Descriptor: | GLYCEROL, SULFATE ION, UvrABC system protein C | | Authors: | Karakas, E, Truglio, J.J, Kisker, C. | | Deposit date: | 2006-11-02 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
5L93
 
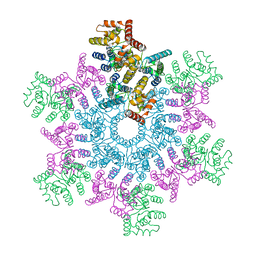 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
2O1T
 
 | |
2NRT
 
 | |
6I3H
 
 | | Crystal structure of influenza A virus M1 N-terminal domain (G18A mutation) | | Descriptor: | Matrix protein 1, PHOSPHATE ION | | Authors: | Miyake, Y, Keusch, J.J, Decamps, L, Ho-Xuan, H, Iketani, S, Gut, H, Kutay, U, Helenius, A, Yamauchi, Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Nat Microbiol, 4, 2019
|
|
2NRW
 
 | |
2FYY
 
 | |
8OJ5
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
2O8O
 
 | |
2G82
 
 | |
2FZ3
 
 | |
2O1U
 
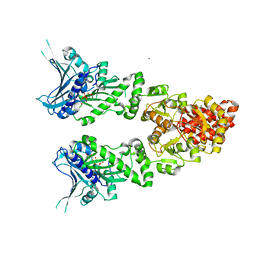 | | Structure of full length GRP94 with AMP-PNP bound | | Descriptor: | Endoplasmin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dollins, D.E, Warren, J.J, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of GRP94-Nucleotide Complexes Reveal Mechanistic Differences between the hsp90 Chaperones.
Mol.Cell, 28, 2007
|
|
2O1V
 
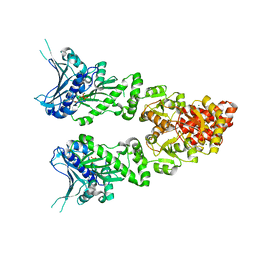 | | Structure of full length GRP94 with ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION | | Authors: | Dollins, D.E, Warren, J.J, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of GRP94-Nucleotide Complexes Reveal Mechanistic Differences between the hsp90 Chaperones.
Mol.Cell, 28, 2007
|
|
2O8C
 
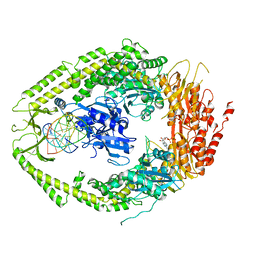 | | human MutSalpha (MSH2/MSH6) bound to ADP and an O6-methyl-guanine T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*(6OG)P*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2J6G
 
 | | FaeG from F4ac ETEC strain 5_95, produced in tobacco plant chloroplast | | Descriptor: | ACETATE ION, FAEG | | Authors: | Van Molle, I, Joensuu, J.J, Buts, L, Panjikar, S, Kotiaho, M, Bouckaert, J, Wyns, L, Niklander-Teeri, V, De Greve, H. | | Deposit date: | 2006-09-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
J.Mol.Biol., 368, 2007
|
|
2N70
 
 | | Two-fold symmetric structure of the 18-60 construct of S31N M2 from Influenza A in lipid bilayers | | Descriptor: | Matrix protein 2 | | Authors: | Andreas, L.B, Reese, M, Eddy, M.T, Gelev, V, Ni, Q, Miller, E.A, Emsley, L, Pintacuda, G, Chou, J.J, Griffin, R.G. | | Deposit date: | 2015-09-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Mechanism of the Influenza A M218-60 Dimer of Dimers.
J.Am.Chem.Soc., 137, 2015
|
|
5JJ8
 
 | | Crystal Structure of the Beta Carbonic Anhydrase psCA3 isolated from Pseudomonas aeruginosa - alternate crystal packing form | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Pinard, M.A, Kurian, J.J, Aggarwal, M, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Cryoannealing-induced space-group transition of crystals of the carbonic anhydrase psCA3.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2NPR
 
 | | Structural Studies on Plasmodium vivax Merozoite Surface Protein-1 | | Descriptor: | Merozoite surface protein 1 | | Authors: | Babon, J.J, Morgan, W.D, Kelly, G, Eccleston, J.F, Feeney, J, Holder, A.A. | | Deposit date: | 2006-10-29 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies on Plasmodium vivax merozoite surface protein-1
Mol.Biochem.Parasitol., 153, 2007
|
|
