4FEN
 
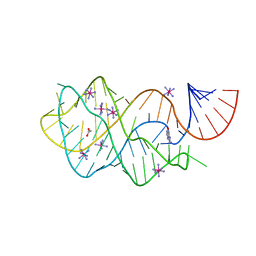 | | Crystal structure of the A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U/U25A/A46G mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
2FS4
 
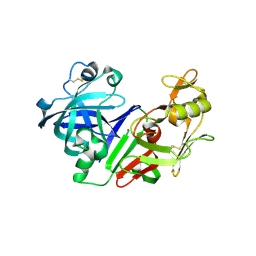 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C ring | | Descriptor: | (6R)-6-({[1-(3-HYDROXYPROPYL)-1,7-DIHYDROQUINOLIN-7-YL]OXY}METHYL)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)PIPERAZIN-2-ONE, Renin | | Authors: | Holsworth, D.D, Cai, C, Cheng, X.-M, Cody, W.L, Downing, D.M, Erasga, N, Lee, C, Powell, N.A, Edmunds, J.J, Stier, M, Jalaie, M, Zhang, E, McConnell, P, Ryan, M.J, Bryant, J, Li, T, Kasani, A, Hall, E, Subedi, R, Rahim, M, Maiti, S. | | Deposit date: | 2006-01-20 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
8IBX
 
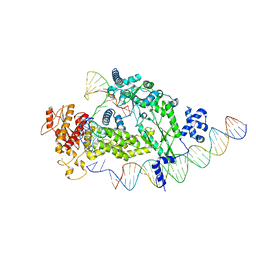 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
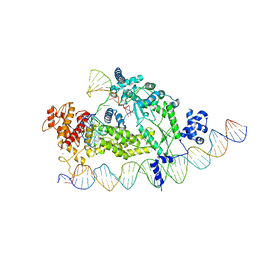 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
4GL9
 
 | | Crystal structure of inhibitory protein SOCS3 in complex with JAK2 kinase domain and fragment of GP130 intracellular domain | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Interleukin-6 receptor subunit beta, PHOSPHATE ION, ... | | Authors: | Kershaw, N.J, Murphy, J.M, Laktyushin, A, Nicola, N.A, Babon, J.J. | | Deposit date: | 2012-08-14 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GOY
 
 | | The crystal structure of human fascin 1 K41A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
6A95
 
 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with tetrodotoxin and Dc1a | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
6AHC
 
 | |
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
4FEP
 
 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
2GE5
 
 | |
4FEO
 
 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
8G1M
 
 | |
8G1L
 
 | |
7CRW
 
 | | Cryo-EM structure of rNLRP1-rDPP9 complex | | Descriptor: | Dipeptidyl peptidase 9, NLR family protein 1 | | Authors: | Huang, M.H, Zhang, X.X, Wang, J, Chai, J.J. | | Deposit date: | 2020-08-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural and biochemical mechanisms of NLRP1 inhibition by DPP9.
Nature, 592, 2021
|
|
7CRV
 
 | |
4E3U
 
 | |
4E37
 
 | | Crystal Structure of P. aeruginosa catalase, KatA tetramer | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | VanderWielen, B.D, Wilson, J.J, Kovall, R.A. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | KatA, the Major Catalase in Pseudomonas aeruginosa, Assists in Protecting Anaerobic Bacteria From Metabolic and Exogenous Nitric Oxide
To be Published
|
|
5XAH
 
 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
4FUP
 
 | |
4G84
 
 | | Crystal structure of human HisRS | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic, ... | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
