1YD0
 
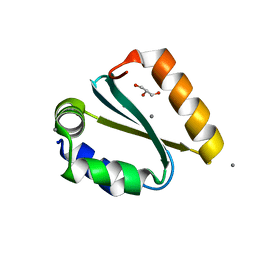 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima bound to its catalytic divalent cation: manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
8DQ2
 
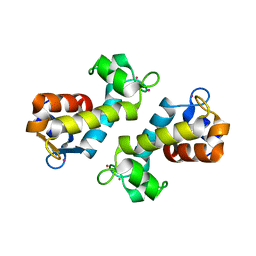 | | X-ray crystal structure of Hansschlegelia quercus lanmodulin (LanM) with lanthanum (III) bound at pH 7 | | Descriptor: | CITRIC ACID, EF-hand domain-containing protein, LANTHANUM (III) ION, ... | | Authors: | Jung, J.J, Lin, C.-Y, Boal, A.K. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced rare-earth separation with a metal-sensitive lanmodulin dimer.
Nature, 618, 2023
|
|
1YD3
 
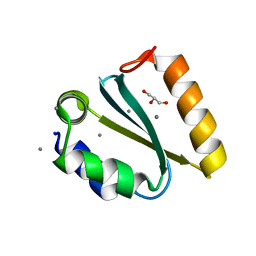 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y43F bound to its catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
1A35
 
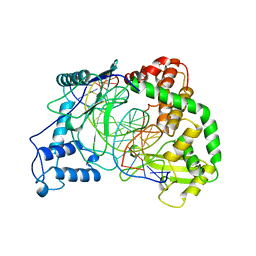 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1YD1
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima bound to its catalytic divalent cation: magnesium | | Descriptor: | GLYCEROL, MAGNESIUM ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
1AIY
 
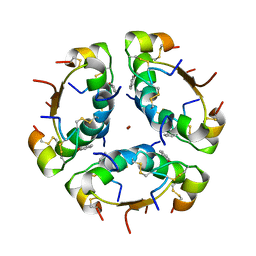 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
4ZUK
 
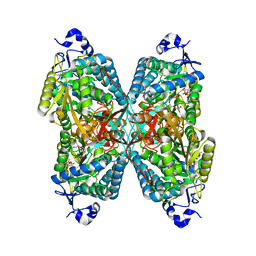 | | Structure ALDH7A1 complexed with NAD+ | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
4I50
 
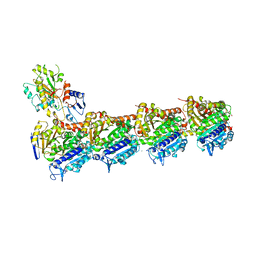 | | Crystal structure of tubulin-stathmin-TTL-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Prota, A.E, Bargsten, K, Zurwerra, D, Field, J.J, Diaz, J.F, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents.
Science, 339, 2013
|
|
1Z71
 
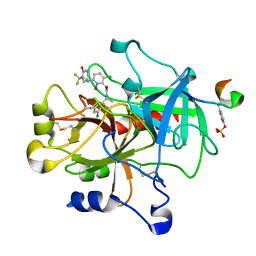 | | thrombin and P2 pyridine N-oxide inhibitor complex structure | | Descriptor: | Hirudin IIIB', L17, thrombin | | Authors: | Nantermet, P.G, Burgey, C.S, Robinson, K.A, Pellicore, J.M, Newton, C.L, Deng, J.Z, Lyle, T.A, Selnick, H.G, Lewis, S.D, Lucas, B.J, Krueger, J.A, Miller-Stein, C, White, R.B, Wong, B, McMasters, D.R, Wallace, A.A, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P(2) pyridine N-oxide thrombin inhibitors: a novel peptidomimetic scaffold
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
6NTH
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by (S) Stereoisomer of A-232 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
4I76
 
 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
3TFK
 
 | | 42F3-p4B10/H2-Ld | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3FSU
 
 | | Crystal Structure of Escherichia coli Methylenetetrahydrofolate Reductase Double Mutant Phe223LeuGlu28Gln complexed with methyltetrahydrofolate | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli.
Biochemistry, 48, 2009
|
|
1ZHK
 
 | | Crystal structure of HLA-B*3501 presenting 13-mer EBV antigen LPEPLPQGQLTAY | | Descriptor: | Beta-2-microglobulin, EBV-peptide LPEPLPQGQLTAY, HLA class I histocompatibility antigen, ... | | Authors: | Tynan, F.E, Borg, N.A, Miles, J.J, Beddoe, T, El-Hassen, D, Silins, S.L, van Zuylen, W.J, Purcell, A.W, Kjer-Nielsen, L, McCluskey, J, Burrows, S.R, Rossjohn, J. | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-17 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structures of highly bulged viral epitopes bound to the major histocompatability class I: Implications for T-cell receptor engagement and T-cell immunodominance
J.Biol.Chem., 280, 2005
|
|
3TF7
 
 | | 42F3 QL9/H2-Ld complex | | Descriptor: | 42F3 Mut7 scFv (42F3 alpha chain, linker, 42F3 beta chain), ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
4JNY
 
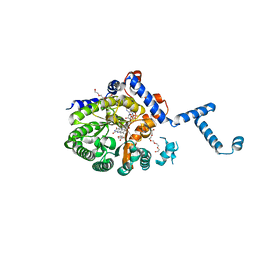 | |
1ZKQ
 
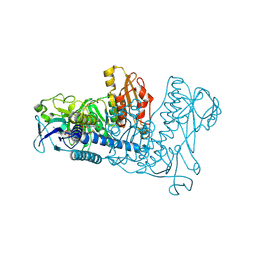 | | Crystal structure of mouse thioredoxin reductase type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 2, mitochondrial | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-05-03 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6MVT
 
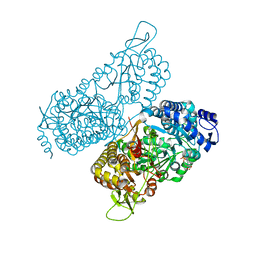 | | Structure of a bacterial ALDH16 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase, SODIUM ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
1D7K
 
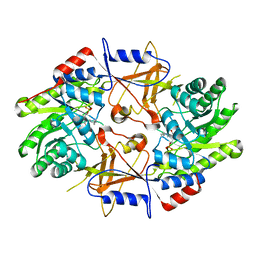 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
4ZUL
 
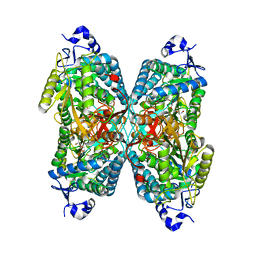 | | Structure ALDH7A1 complexed with alpha-aminoadipate | | Descriptor: | 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase, TETRAETHYLENE GLYCOL, ... | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
1D4E
 
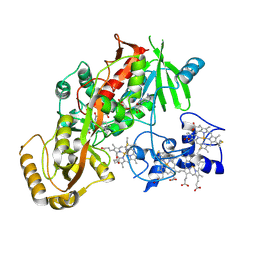 | | CRYSTAL STRUCTURE OF THE FLAVOCYTOCHROME C FUMARATE REDUCTASE OF SHEWANELLA PUTREFACIENS STRAIN MR-1 COMPLEXED WITH FUMARATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C FUMARATE REDUCTASE, FUMARIC ACID, ... | | Authors: | Leys, D, Tsapin, A.S, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 1999-10-03 | | Release date: | 1999-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1.
Nat.Struct.Biol., 6, 1999
|
|
5CU9
 
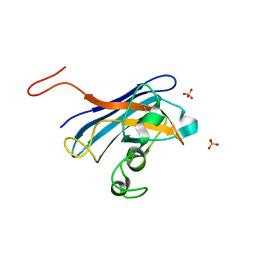 | | CANDIDA ALBICANS SUPEROXIDE DISMUTASE 5 (SOD5), APO | | Descriptor: | Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Waninger-Saroni, J.J, Taylor, A.B, Hart, P.J, Galaleldeen, A. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | CANDIDA ALBICANS SUPEROXIDE DISMUTASE 5 (SOD5), APO
To Be Published
|
|
3DG1
 
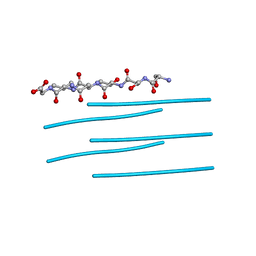 | | Segment SSTNVG derived from IAPP | | Descriptor: | SSTNVG from Islet Amyloid Polypeptide | | Authors: | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
1YD5
 
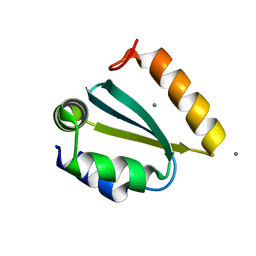 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant N88A bound to its catalytic divalent cation | | Descriptor: | MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
1YD4
 
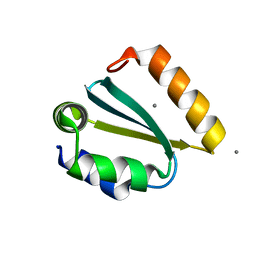 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y29F bound to its catalytic divalent cation | | Descriptor: | MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
