6RYD
 
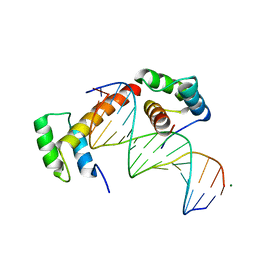 | | WUS-HD bound to TGAA DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*CP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYL
 
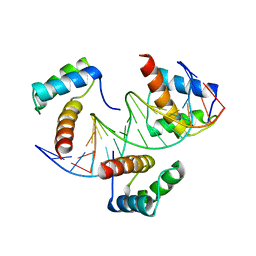 | | WUS-HD bound to TAAT DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*AP*AP*CP*CP*CP*AP*TP*TP*AP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*TP*AP*AP*TP*GP*GP*GP*TP*TP*GP*TP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6WU1
 
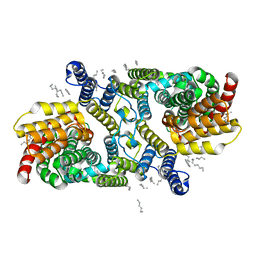 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6X3V
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus etomidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Etomidate, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
6VJV
 
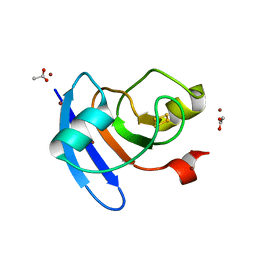 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
6X0K
 
 | |
6X3T
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
6WH7
 
 | | Capsid structure of Penaeus monodon metallodensovirus following EDTA treatment | | Descriptor: | Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1CIL
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
6V0Z
 
 | | Structure of ALDH7A1 mutant R441C complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Korasick, D.A, Tanner, J.J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical, structural, and computational analyses of two new clinically identified missense mutations of ALDH7A1.
Chem.Biol.Interact., 2024
|
|
6X3W
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus phenobarbital | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ethyl-5-phenylpyrimidine-2,4,6(1H,3H,5H)-trione, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
6WU2
 
 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
1DW0
 
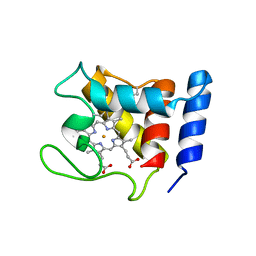 | | STRUCTURE OF OXIDIZED SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
6WTW
 
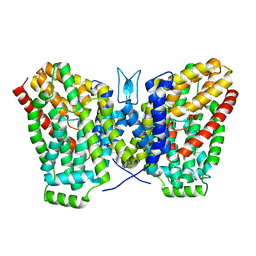 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
1DW1
 
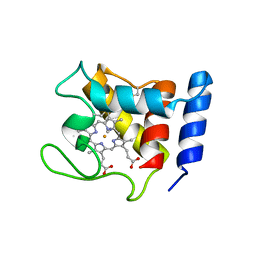 | | STRUCTURE OF THE CYANIDE COMPLEX OF SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
6WJ9
 
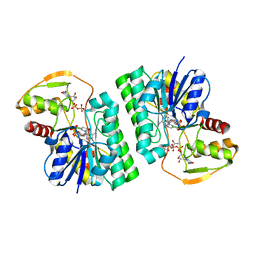 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WU4
 
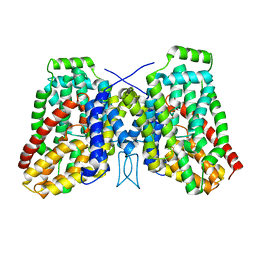 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
1DW2
 
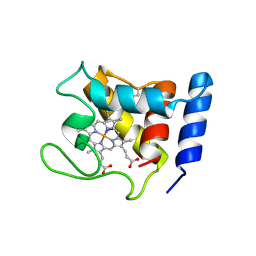 | | STRUCTURE OF THE NITRIC OXIDE COMPLEX OF REDUCED SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
1DW3
 
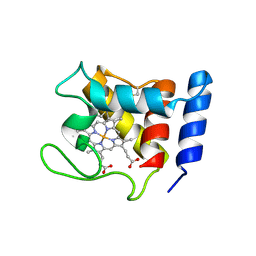 | | STRUCTURE OF A REDUCED OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
1G37
 
 | | CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN COMPLEXED WITH BCH-10556 AND EXOSITE-DIRECTED PEPTIDE | | Descriptor: | 3-(4-AMINO-CYCLOHEXYL)-2-HYDROXY-3-[(4-OXO-2-PHENYLMETHANESULFONYL-1,2,3,4-TETRAHYDRO-PYRROLO[1,2-A]PYRAZINE-6-CARBONYL)-AMINO]-PROPIONIC ACID BUTYL ESTER, ALPHA THROMBIN, THROMBIN NONAPEPTIDE INHIBITOR | | Authors: | Bachand, B, Tarazi, M, St-Denis, Y, Edmunds, J.J, Winocour, P.D, Leblond, L, Siddiqui, M.A. | | Deposit date: | 2000-10-23 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective bicyclic lactam inhibitors of thrombin. Part 4: transition state inhibitors.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1EO4
 
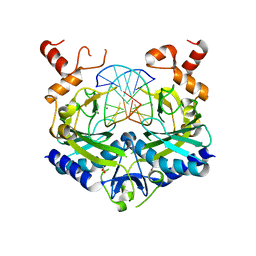 | | ECORV BOUND TO MN2+ AND COGNATE DNA CONTAINING A 3'S SUBSTITION AT THE CLEAVAGE SITE | | Descriptor: | ACETIC ACID, DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
1EOO
 
 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EUY
 
 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA MUTANT AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-19 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
1GMY
 
 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
