4JXZ
 
 | |
5NG1
 
 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
4JZT
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8VVM
 
 | | Structure of FabS1CE1-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7QLL
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLK
 
 | |
8VTR
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8VU4
 
 | | Structure of FabS1CE4-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7Q6T
 
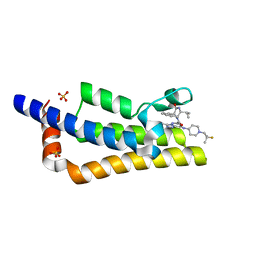 | | Crystal structure of the bromodomain of ATAD2 with AZ13824374 | | Descriptor: | (1R,9S,12R)-13-[[8-[[1-(2-fluoranyl-2-methyl-propyl)piperidin-4-yl]amino]-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-12-propan-2-yl-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
4JYZ
 
 | |
7Q6W
 
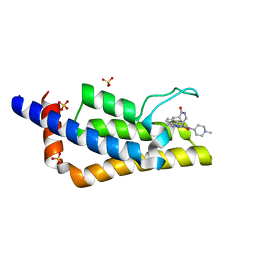 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridazine (cpd 22) | | Descriptor: | (1R,9S)-13-[[3-methyl-8-[(1-methylpiperidin-4-yl)amino]-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6U
 
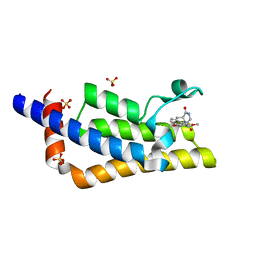 | | Crystal structure of the bromodomain of ATAD2 with phenol HTS hit (cpd 6) | | Descriptor: | (1R,9S)-13-(3,5-dimethoxy-4-oxidanyl-phenyl)carbonyl-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6V
 
 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridine (cpd 14) | | Descriptor: | (1R,9S)-13-[(8-azanyl-3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
8VTP
 
 | | Structure of FabS1CE-EPR-1, a high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VU1
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (trigonal form) | | Descriptor: | S1CE3 VARIANT OF FAB-EPR-1 heavy chain, S1CE3 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
2V5C
 
 | | Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure | | Descriptor: | CACODYLATE ION, CALCIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2VJJ
 
 | | TAILSPIKE PROTEIN OF E.COLI BACTERIOPHAGE HK620 IN COMPLEX WITH HEXASACCHARIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mueller, J.J, Barbirz, S, Uetrecht, C, Seckler, R, Heinemann, U. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Escherichia Coli Phage Hk620 Tailspike: Podoviral Tailspike Endoglycosidase Modules are Evolutionarily Related.
Mol.Microbiol., 69, 2008
|
|
1RV0
 
 | | 1930 Swine H1 Hemagglutinin complexed with LSTA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose, hemagglutinin | | Authors: | Skehel, J.J, Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
8FX9
 
 | |
6WAN
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
7SQN
 
 | |
