4GWA
 
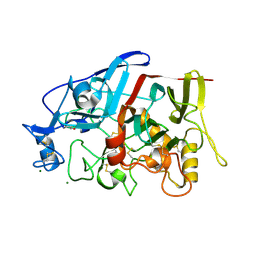 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5EEV
 
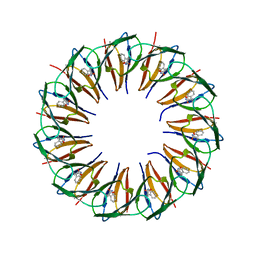 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 3.88 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2UZS
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2UWD
 
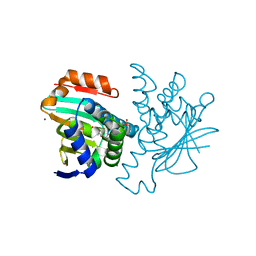 | | Inhibition of the HSP90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole, isoxazole amide analogs | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION, ... | | Authors: | Sharp, S.Y, Prodromou, C, Boxall, K, Powers, M.V, Holmes, J.L, Box, G, Matthews, T.P, Cheung, K.M, Kalusa, A, James, K, Hayes, A, Hardcastle, A, Dymock, B, Brough, P.A, Barril, X, Cansfield, J.E, Wright, L.M, Surgenor, A, Foloppe, N, Aherne, W, Pearl, L, Jones, K, McDonald, E, Raynaud, F, Eccles, S, Drysdale, M, Workman, P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of the heat shock protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues.
Mol. Cancer Ther., 6, 2007
|
|
6DHU
 
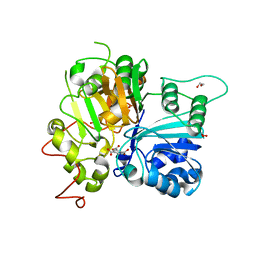 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT0911 from cocktail soak | | Descriptor: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, benzene-1,2,4-tricarboxylic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
2UVD
 
 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
6DIE
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment benzene-1,2,4-tricarboxylic acid from single soak | | Descriptor: | 1,2-ETHANEDIOL, Tdp1 catalytic domain (residues 149-609), benzene-1,2,4-tricarboxylic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJF
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ502 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyquinoline-3,8-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DJE
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound CDS010292 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-(propan-2-yl)quinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
1FHZ
 
 | | PSORALEN CROSS-LINKED D(CCGGTACCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
1S2O
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | | Descriptor: | MAGNESIUM ION, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.L. | | Deposit date: | 2004-01-09 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1FHY
 
 | | PSORALEN CROSS-LINKED D(CCGCTAGCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, CALCIUM ION, DNA (5'-D(*CP*CP*GP*CP*TP*AP*GP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
4FRN
 
 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
2W9Z
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W9F
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-23 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4GRH
 
 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
2WGP
 
 | | Crystal structure of human dual specificity phosphatase 14 | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 14, PHOSPHATE ION | | Authors: | Lountos, G.T, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Overproduction, Purification and Structure Determination of Human Dual-Specificity Phosphatase 14.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1RRK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RS0
 
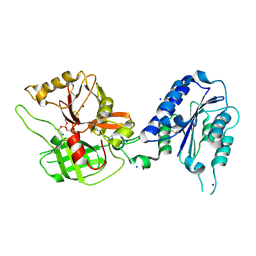 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1FLZ
 
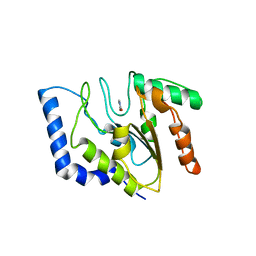 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
5DA7
 
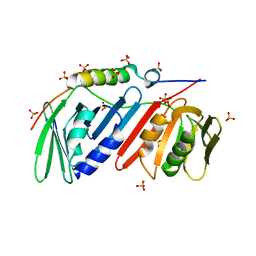 | | monomeric PCNA bound to a small protein inhibitor | | Descriptor: | DNA polymerase sliding clamp 1, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Ladner, J.E, Altieri, A.S, Kelman, Z. | | Deposit date: | 2015-08-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Nucleic Acids Res., 44, 2016
|
|
2WAG
 
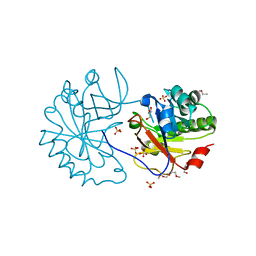 | | The Structure of a family 25 Glycosyl hydrolase from Bacillus anthracis. | | Descriptor: | GLYCEROL, LYSOZYME, PUTATIVE, ... | | Authors: | Martinez-Fleites, C, Korczynska, J.E, Cope, M, Turkenburg, J.P, Taylor, E.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of a Family Gh25 Lysozyme from Bacillus Anthracis Implies a Neighboring-Group Catalytic Mechanism with Retention of Anomeric Configuration
Carbohydr.Res., 344, 2009
|
|
2W1E
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(PHENYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-5-YL)METHYL]MORPHOLIN-4-IUM, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
