3CQS
 
 | | A 3'-OH, 2',5'-phosphodiester substitution in the hairpin ribozyme active site reveals similarities with protein ribonucleases | | 分子名称: | 13-mer substrate strand with 3'-OH, 2',5'-phosphodiester covalently linking 5th and 6th nucleotides, 19-mer ribozyme strand, ... | | 著者 | Torelli, A.T, Spitale, R.C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2008-04-03 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme
Biochem.Biophys.Res.Commun., 371, 2008
|
|
8DQ1
 
 | | Quorum-sensing receptor RhlR bound to PqsE | | 分子名称: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, DNA (5'-D(*AP*CP*CP*TP*GP*CP*CP*AP*GP*AP*CP*TP*GP*CP*AP*CP*AP*G)-3'), ... | | 著者 | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | 登録日 | 2022-07-18 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | 分子名称: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | 著者 | Zahn, M, McGeehan, J.E. | | 登録日 | 2021-05-25 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | 分子名称: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
3I2B
 
 | | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase | | 分子名称: | 1,2-ETHANEDIOL, 6-pyruvoyl tetrahydrobiopterin synthase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ugochukwu, E, Cocking, R, Pilka, E, Yue, W.W, Bray, J.E, Chaikuad, A, Krojer, T, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2009-06-29 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase
To be Published
|
|
8G7M
 
 | | ATP-bound mtHsp60 V72I focus | | 分子名称: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | 分子名称: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
1MR2
 
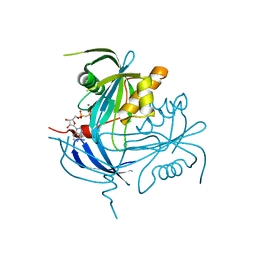 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | 分子名称: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | 著者 | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | 登録日 | 2002-09-17 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
8G7N
 
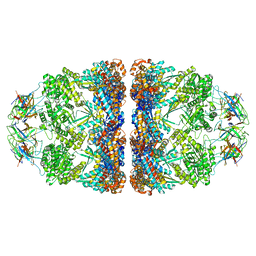 | | ATP- and mtHsp10-bound mtHsp60 V72I | | 分子名称: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | 分子名称: | 60 kDa heat shock protein, mitochondrial | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
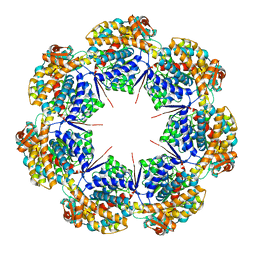 | | mtHsp60 V72I apo focus | | 分子名称: | 60 kDa heat shock protein, mitochondrial | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
3ZR5
 
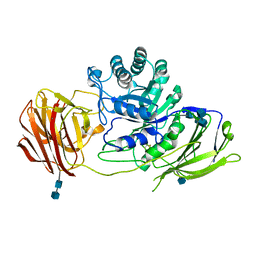 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | 登録日 | 2011-06-14 | | 公開日 | 2011-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3JWO
 
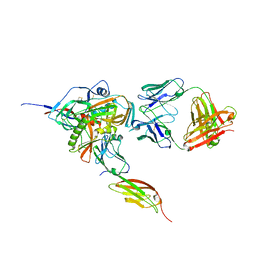 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | 著者 | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | 登録日 | 2009-09-18 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ZFM
 
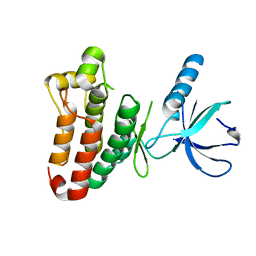 | | Crystal structure of EphB2 | | 分子名称: | EPHRIN TYPE-B RECEPTOR 2 | | 著者 | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | 登録日 | 2012-12-12 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZEW
 
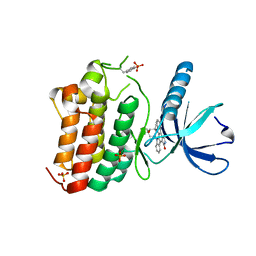 | | Crystal structure of EphB4 in complex with staurosporine | | 分子名称: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE, SULFATE ION | | 著者 | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | 登録日 | 2012-12-07 | | 公開日 | 2013-12-25 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3JWD
 
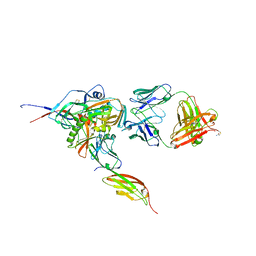 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | 著者 | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | 登録日 | 2009-09-18 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ZIA
 
 | | The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Robinson, G.C, Bason, J.V, Montgomery, M.G, Fearnley, I.M, Mueller, D.M, Leslie, A.G.W, Walker, J.E. | | 登録日 | 2013-01-07 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structure of F1-ATPase from Saccharomyces Cerevisiae Inhibited by its Regulatory Protein If1.
Open Biol., 3, 2013
|
|
8RIW
 
 | | T2R-TTL-1-L01 complex | | 分子名称: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | 登録日 | 2023-12-19 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
3ZTJ
 
 | | Structure of influenza A neutralizing antibody selected from cultures of single human plasma cells in complex with human H3 Influenza haemagglutinin. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY HEAVY CHAIN, ... | | 著者 | Voss, J.E, Vachieri, S.G, Gamblin, S.J, Collins, P.J, Haire, L.F, Skehel, J.J. | | 登録日 | 2011-07-08 | | 公開日 | 2011-08-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
1OKH
 
 | | Viscotoxin A3 from Viscum album L. | | 分子名称: | PHOSPHATE ION, SULFATE ION, VISCOTOXIN A3 | | 著者 | Debreczeni, J.E, Girmann, B, Zeeck, A, Sheldrick, G.M. | | 登録日 | 2003-07-24 | | 公開日 | 2003-12-04 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of Viscotoxin A3: Disulfide Location from Weak Sad Data
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3ZLC
 
 | | Crystal Structure of Erv41p | | 分子名称: | ER-DERIVED VESICLES PROTEIN ERV41 | | 著者 | Biterova, E.I, Svard, M, Possner, D.D.D, Guy, J.E. | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2013-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | The Crystal Structure of the Lumenal Domain of Erv41P, a Protein Involved in Transport between the Endoplasmic Reticulum and Golgi Apparatus
J.Mol.Biol., 425, 2013
|
|
3ZGP
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | 分子名称: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | 著者 | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | 登録日 | 2012-12-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZTN
 
 | | STRUCTURE OF INFLUENZA A NEUTRALIZING ANTIBODY SELECTED FROM CULTURES OF SINGLE HUMAN PLASMA CELLS IN COMPLEX WITH HUMAN H1 INFLUENZA HAEMAGGLUTININ. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY LIGHT CHAIN, ... | | 著者 | Hubbard, P.A, Ritchie, A.J, Corti, D, Voss, J.E, Gamblin, S.J, Codoni, G, Macagno, A, Jarrossay, D, Pinna, D, Minola, A, Vanzetta, F, Silacci, C, Fernandez-Rodriguez, B.M, Agatic, G, Giacchetto-Sasselli, I, Vachieri, S.G, Sallusto, F, Collins, P.J, Haire, L.F, Temperton, N, Langedijk, J.P.M, Skehel, J.J, Lanzavecchia, A. | | 登録日 | 2011-07-12 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
6DU3
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | 分子名称: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION, REST-pS861 | | 著者 | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | 登録日 | 2018-06-19 | | 公開日 | 2018-09-26 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
3DSE
 
 | |
