2YEX
 
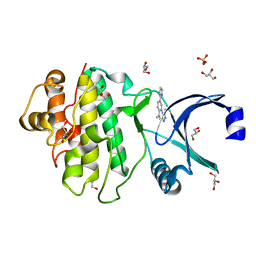 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 5-METHYL-8-(1H-PYRROL-2-YL)[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BZD
 
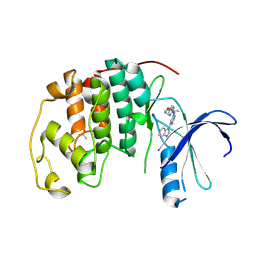 | |
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
1WWJ
 
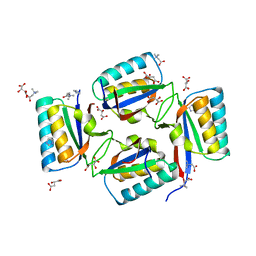 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D0S
 
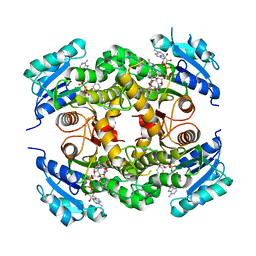 | | Mtb InhA complex with Pyradizinone compound 14 | | Descriptor: | 1-{4-[(acetylamino)methyl]phenyl}-4-(4-chlorophenoxy)-6-oxo-1,6-dihydropyridazine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], MAGNESIUM ION, ... | | Authors: | Read, J.A, Breed, J, Madhavapeddi, P, Lange, S. | | Deposit date: | 2014-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyridazinones: A Novel Scaffold with Excellent Physicochemical Properties and Safety Profile for a Clinically Validated Target of Mycobacterium Tuberculosis
To be Published
|
|
4COS
 
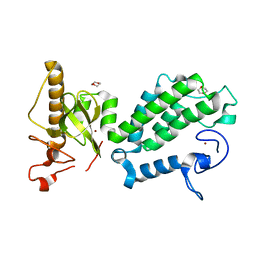 | | Crystal structure of the PHD-Bromo-PWWP cassette of human PRKCBP1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PROTEIN KINASE C-BINDING PROTEIN 1, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Newman, J.A, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Filippakopoulos, P. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin.
Cell Rep, 17, 2016
|
|
4CRS
 
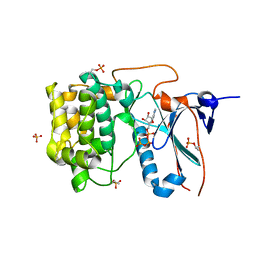 | | Human Protein Kinase N2 (PKN2, PRKCL2) in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SERINE/THREONINE-PROTEIN KINASE N2, ... | | Authors: | Mathea, S, Elkins, J.M, Shrestha, L, Szklarz, M, Tallant, C, Newman, J.A, Cooper, C.D, Shrestha, B, Tumber, A, Cocking, R, Salah, E, von Delft, F, Arrowsmith, C, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Pkn2
To be Published
|
|
1WUE
 
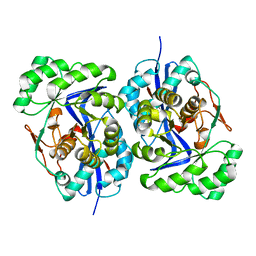 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
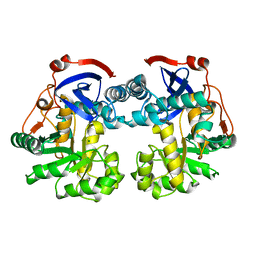 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1BJK
 
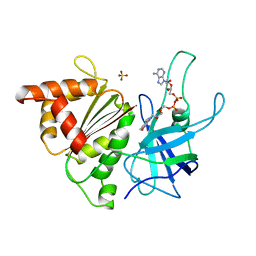 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
4D0R
 
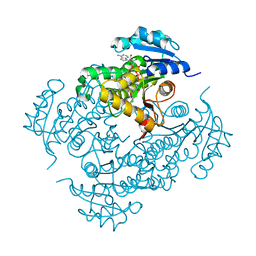 | | Mtb InhA complex with Pyradizinone compound 1 | | Descriptor: | 4-(4-chlorophenoxy)-6-oxo-1-phenyl-1,6-dihydropyridazine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Lange, S. | | Deposit date: | 2014-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyridazinones: A Novel Scaffold with Excellent Physicochemical Properties and Safety Profile for a Clinically Validated Target of Mycobacterium Tuberculosis
To be Published
|
|
1W8C
 
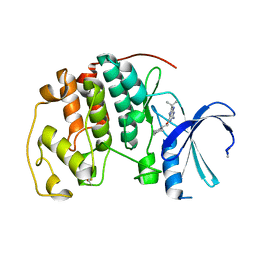 | |
4CK0
 
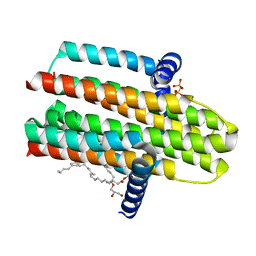 | | Crystal structure of the integral membrane diacylglycerol kinase - form 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIACYLGLYCEROL KINASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Li, D, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2013-12-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase with Zn-Amppcp Bound and its Catalytic Mechanism
To be Published
|
|
1Y7M
 
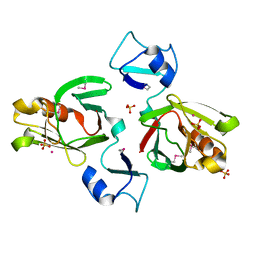 | | Crystal Structure of the B. subtilis YkuD protein at 2 A resolution | | Descriptor: | CADMIUM ION, SULFATE ION, hypothetical protein BSU14040 | | Authors: | Bielnicki, J.A, Devedjiev, Y, Derewenda, U, Dauter, Z, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes.
Proteins, 62, 2006
|
|
4DHD
 
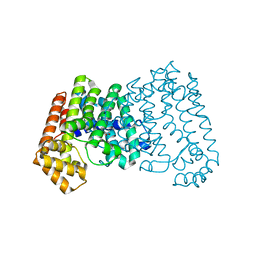 | | Crystal structure of isoprenoid synthase A3MSH1 (TARGET EFI-501992) from Pyrobaculum calidifontis | | Descriptor: | ACETATE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1BQE
 
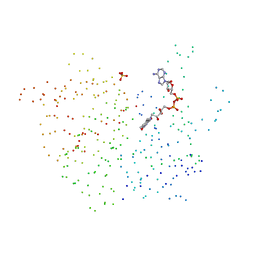 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY (T155G) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-08-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis.
J.Biol.Chem., 276, 2001
|
|
1QCY
 
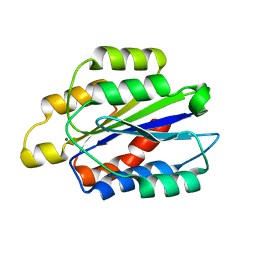 | | THE CRYSTAL STRUCTURE OF THE I-DOMAIN OF HUMAN INTEGRIN ALPHA1BETA1 | | Descriptor: | I-DOMAIN OF INTEGRIN ALPHA1BETA1, MAGNESIUM ION | | Authors: | Kankare, J.A, Salminen, T.A, Nymalm, Y, Kaepylae, J, Heino, J, Johnson, M.S. | | Deposit date: | 1999-05-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Jararhagin-derived RKKH Peptides Induce Structural Changes in a1I Domain of Human Integrin a1b1
J.Biol.Chem., 279, 2004
|
|
1A2Z
 
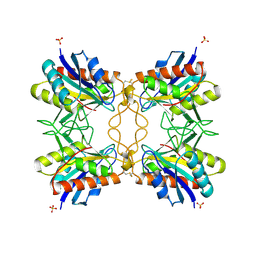 | |
4C1W
 
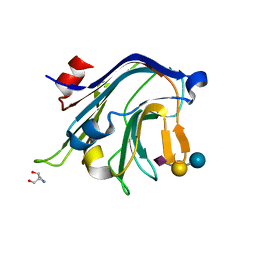 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, NEURAMINIDASE | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Prevention of Influenza by Targeting Host Receptors Using Engineered Proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1Z09
 
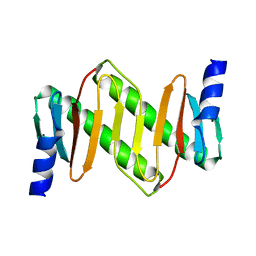 | | Solution structure of km23 | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Ilangovan, U, Ding, W, Mulder, K, Hinck, A.P, Zuniga, J, Trbovich, J.A, Demeler, B, Groppe, J.C. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Homodimeric Dynein Light Chain km23.
J.Mol.Biol., 352 (2), 2005
|
|
5T4U
 
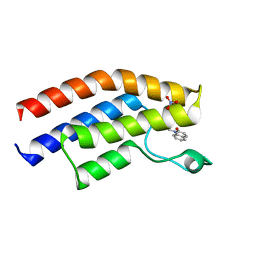 | | Crystal structure of the bromodomain of human BRPF1 in complex with a quinolinone ligand | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, NITRATE ION, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
5UC3
 
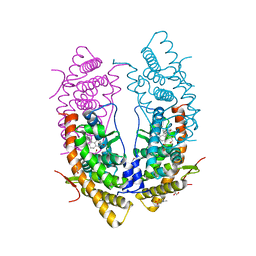 | | Structure of the dominant negative mutant Glucocorticoid Receptor alpha (L733K/N734P) complexed with RU-486 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, ... | | Authors: | Min, J, Cidlowski, J.A, Pedersen, L.C. | | Deposit date: | 2016-12-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Nuclear receptor
To Be Published
|
|
5T4V
 
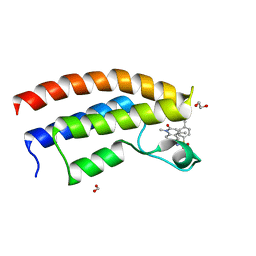 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-48 ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-N-(7-methoxy-1,4-dimethyl-2-oxo-1,2-dihydroquinolin-6-yl)benzene-1-sulfonamide, FORMIC ACID, ... | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
5SVH
 
 | |
