6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
1WAK
 
 | | X-ray structure of SRPK1 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE SPRK1 | | Authors: | Ngo, J.C, Gullingsrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.D, Adams, J.A, Mccammon, J.A, Ghosh, G. | | Deposit date: | 2004-10-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Sr Protein Kinase 1 is Resilient to Inactivation.
Structure, 15, 2007
|
|
1MU8
 
 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
1MUE
 
 | | Thrombin-Hirugen-L405,426 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDO-2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUOROPHENYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Nantermet, P.G, Selnick, H.G, Isaacs, R.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Stranieri, M.T, Cook, J.J, McMasters, D.R, Pellicore, J.M, Pal, S, Wallace, A.A, Clayton, F.C, Bohn, D, Welsh, D.C, Lynch, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacokinetic optimization of 3-amino-6-chloropyrazinone acetamide thrombin inhibitors. Implementation of P3 pyridine N-oxides to deliver an orally bioavailable series containing P1 N-benzylamides.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1MU6
 
 | | Crystal Structure of Thrombin in Complex with L-378,622 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
6MCC
 
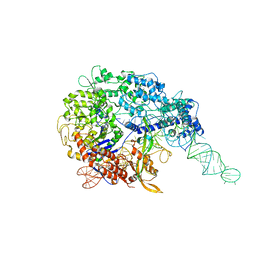 | |
6M9I
 
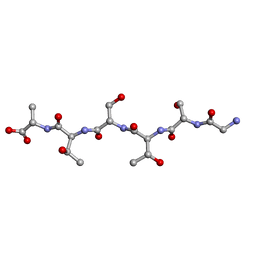 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
4V7S
 
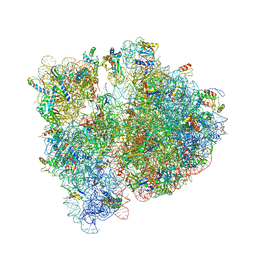 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4US6
 
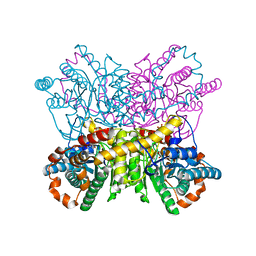 | | New Crystal Form of Glucose Isomerase Grown in Short Peptide Supramolecular Hydrogels | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Diaz-Mochon, J.J, Alvarez de Cienfuegos, L. | | Deposit date: | 2014-07-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Influence of the Chirality of Short Peptide Supramolecular Hydrogels in Protein Crystallogenesis.
Chem.Commun.(Camb.), 51, 2015
|
|
4W2O
 
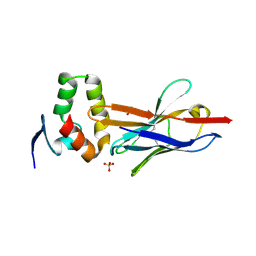 | |
4W2P
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody C | | Descriptor: | ACETATE ION, Anti-Marburgvirus Nucleoprotein Single Domain Antibody C, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
4W2Q
 
 | |
4W4G
 
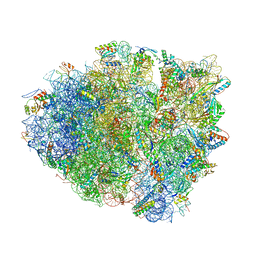 | | Postcleavage state of 70S bound to HigB toxin and AAA (lysine) codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V6D
 
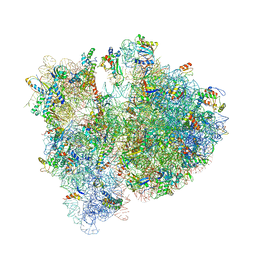 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
1MNS
 
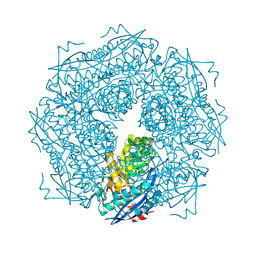 | |
6XTJ
 
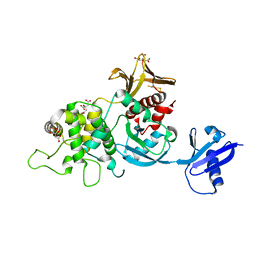 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
6XLQ
 
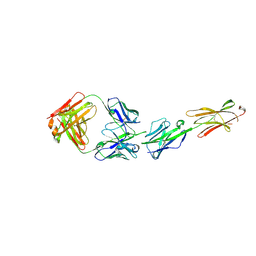 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
4US3
 
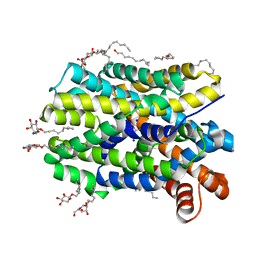 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4US4
 
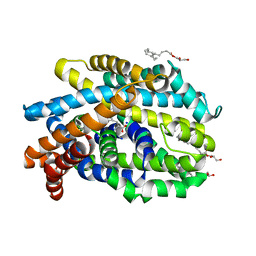 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RIY
 
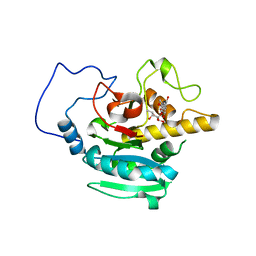 | | B-specific-1,3-galactosyltransferase (GTB)+H-antigen acceptor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ4
 
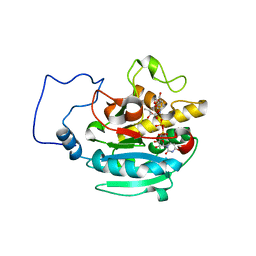 | | B-specific alpha-1,3-galactosyltransferase G176R +UDP+ADA | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-14 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RIX
 
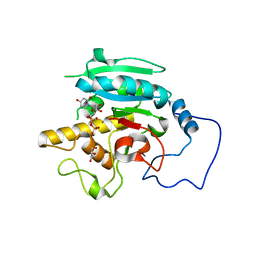 | | B-specific-1,3-galactosyltransferase)(GTB) + UDP | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ9
 
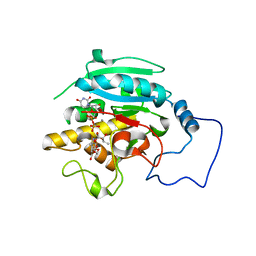 | |
2RJ0
 
 | |
