2JFA
 
 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH AN AFFINITY-SELECTED COREPRESSOR PEPTIDE | | Descriptor: | COREPRESSOR PEPTIDE, ESTROGEN RECEPTOR, RALOXIFENE, ... | | Authors: | Heldring, N, Pawson, T, McDonnell, D, Treuter, E, Gustafsson, J.A, Pike, A.C.W. | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into Corepressor Recognition by Antagonist-Bound Estrogen Receptors.
J.Biol.Chem., 282, 2007
|
|
2JIY
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M149 REPLACED WITH TRP (CHAIN M, AM149W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
2JJ0
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M248 REPLACED WITH TRP (CHAIN M, AM248W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
2JGW
 
 | | Structure of CCP module 7 of complement factor H - The AMD at risk varient (402H) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
2JF9
 
 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH A TAMOXIFEN-SPECIFIC PEPTIDE ANTAGONIST | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYTAMOXIFEN, AB5 PEPTIDE, ... | | Authors: | Heldring, N, Pawson, T, McDonnell, D, Treuter, E, Gustafsson, J.A, Pike, A.C.W. | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into Corepressor Recognition by Antagonist-Bound Estrogen Receptors.
J.Biol.Chem., 282, 2007
|
|
2K31
 
 | | Solution Structure of cGMP-binding GAF domain of Phosphodiesterase 5 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, Phosphodiesterase 5A, cGMP-specific | | Authors: | Heikaus, C.C, Stout, J.R, Sekharan, M.R, Eakin, C.M, Rajagopal, P, Brzovic, P.S, Beavo, J.A, Klevit, R.E. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the cGMP Binding GAF Domain from Phosphodiesterase 5: Insights into Nucleotide Selectivity, Dimerization, and cGMP-Dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
2JQ4
 
 | | Complete resonance assignments and solution structure calculation of ATC2521 (NESG ID: AtT6) from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2571 | | Authors: | Srisailam, S, Lemak, A, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a protein (ATC2521)of unknown function from Agrobacterium tumefaciens
To be Published
|
|
2JN4
 
 | | Solution NMR Structure of Protein RP4601 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium Target RpT2; Ontario Center for Structural Proteomics Target RP4601. | | Descriptor: | Hypothetical protein fixU, nifT | | Authors: | Lemak, A, Srisailam, S, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hypothetical protein RP4601 from Rhodopseudomonas palustris
To be Published
|
|
2LS9
 
 | | Pleurocidin-NH2 | | Descriptor: | Pleurocidin | | Authors: | Vermeer, L.S, Kozlowska, J, Mason, J.A. | | Deposit date: | 2012-04-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | All Atom Simulations of the Initial Binding of Magainin and Pleurocidin to Membranes Comprising of a Mixture of Anionic and Zwitterionic Lipids
To be Published
|
|
2L99
 
 | | Solution structure of LAK160-P10 | | Descriptor: | LAK160-P10 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L9A
 
 | | Solution structure of LAK160-P12 | | Descriptor: | LAK160-P12 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2LKK
 
 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2LAQ
 
 | |
2LFW
 
 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LSA
 
 | | Magainin | | Descriptor: | Magainin-2 | | Authors: | Vermeer, L.S, Mason, J.A. | | Deposit date: | 2012-04-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | All Atom Simulations of the Initial Binding of Magainin and Pleurocidin to Membranes Comprising a Mixture of Anionic and Zwitterionic Lipids
To be Published
|
|
1EH6
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE | | Descriptor: | O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE, ZINC ION | | Authors: | Daniels, D.S, Tainer, J.A. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding.
EMBO J., 19, 2000
|
|
5I34
 
 | | Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP | | Descriptor: | Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, INOSINIC ACID | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
5I66
 
 | | X-ray structure of the Fab fragment of 8B6, a murine monoclonal antibody specific for the human serotonin transporter | | Descriptor: | 8B6 antibody, heavy chain, light chain | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5I75
 
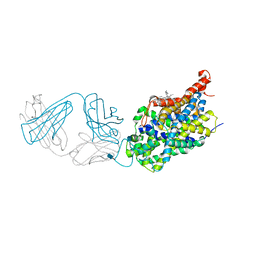 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central site and Br-citalopram at the allosteric site | | Descriptor: | (1S)-1-(4-bromophenyl)-1-[3-(dimethylamino)propyl]-1,3-dihydro-2-benzofuran-5-carbonitrile, (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
1E5N
 
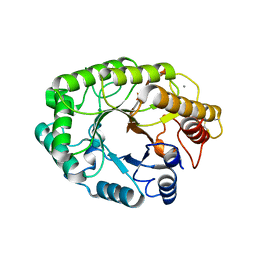 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
1ET6
 
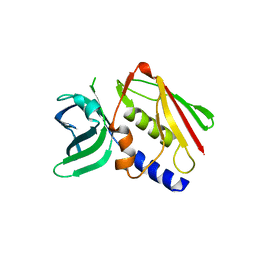 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SMEZ-2 | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EU3
 
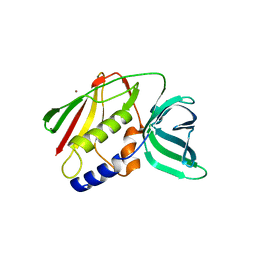 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SUPERANTIGEN SMEZ-2, ... | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EYV
 
 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
5IM2
 
 | | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate | | Descriptor: | BENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate
To be published
|
|
