1OVG
 
 | | M64V PNP +MePdr | | Descriptor: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, P.W, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-26 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
6FCU
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1OU4
 
 | | Native PNP +Talo | | Descriptor: | 6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, W.B, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-24 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
6PAX
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PAX-6 PAIRED DOMAIN-DNA COMPLEX REVEALS A GENERAL MODEL FOR PAX PROTEIN-DNA INTERACTIONS | | Descriptor: | 26 NUCLEOTIDE DNA, HOMEOBOX PROTEIN PAX-6 | | Authors: | Xu, H.E, Rould, M.A, Xu, W, Epstein, J.A, Maas, R.L, Pabo, C.O. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding.
Genes Dev., 13, 1999
|
|
6OZZ
 
 | | N terminally deleted GapR crystal structure from C. crescentus | | Descriptor: | UPF0335 protein CC_3319 | | Authors: | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
6PBK
 
 | |
6ESL
 
 | | Crystal structure of the Legionella pneumoppila LapA | | Descriptor: | Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Richardson, K, Garnett, J.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Type II Secretion-Dependent Aminopeptidase LapA and Acyltransferase PlaC Are Redundant for Nutrient Acquisition duringLegionella pneumophilaIntracellular Infection of Amoebas.
MBio, 9, 2018
|
|
6MMO
 
 | |
6MMQ
 
 | |
6F9G
 
 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
6MRH
 
 | |
1OTX
 
 | | Purine Nucleoside Phosphorylase M64V mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, W.B, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-23 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
6F59
 
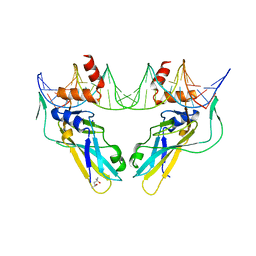 | | Crystal structure of human Brachyury (T) G177D variant in complex with DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Brachyury protein, DNA (26-MER), ... | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Crystal structure of human Brachyury (T) G177D variant in complex with DNA
To Be Published
|
|
6MHB
 
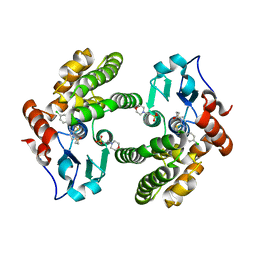 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
1OTY
 
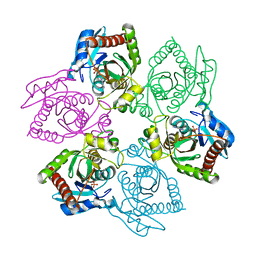 | | Native PNP +ALLO | | Descriptor: | 6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, W.B, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-23 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
6MIE
 
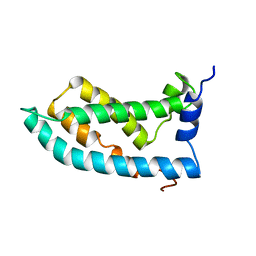 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6MM2
 
 | |
6MMC
 
 | |
6MRE
 
 | |
6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MRC
 
 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
6MRI
 
 | |
6MX5
 
 | |
6OZW
 
 | |
6MI5
 
 | | NMR solution structure of lanmodulin (LanM) complexed with yttrium(III) ions | | Descriptor: | Lanmodulin, YTTRIUM (III) ION | | Authors: | Cook, E.C, Featherson, E.R, Showalter, S.A, Cotruvo Jr, J.A. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Rare Earth Element Recognition by Methylobacterium extorquens Lanmodulin.
Biochemistry, 58, 2019
|
|
