7KTI
 
 | | DNA Polymerase Mu, 8-oxodGTP:Ct Product State Ternary Complex, 20 uM Mn2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTD
 
 | | DNA Polymerase Mu, 8-oxodGTP:Ct Product State Ternary Complex, 10 mM Mn2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTM
 
 | | DNA Polymerase Mu (K438D), 8-oxodGTP:Ct Reaction State Ternary Complex, 50 mM Mn2+ (30min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTJ
 
 | | DNA Polymerase Mu (K438D), 8-oxodGTP:Ct Pre-Catalytic Ground State Ternary Complex, 20 mM Ca2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTL
 
 | | DNA Polymerase Mu (K438D), 8-oxodGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (90min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTK
 
 | | DNA Polymerase Mu (K438D), 8-oxodGTP:Ct Ground State Ternary Complex, 50 mM Mg2+ (90min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
4RQK
 
 | |
4RK2
 
 | | Crystal structure of sugar transporter RHE_PF00321 from Rhizobium etli, target EFI-510806, an open conformation | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative sugar ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sugar Transporter RHE_Pf00321 from Rhizobium Etli, Target EFI-510806
To be Published
|
|
4RKR
 
 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007, complex with lactose | | Descriptor: | Transcriptional regulator, LacI family, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
3QKU
 
 | | Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4RXT
 
 | | Crystal structure of carbohydrate transporter solute binding protein Arad_9553 from Agrobacterium Radiobacter, Target EFI-511541, in complex with D-arabinose | | Descriptor: | Sugar ABC transporter, alpha-L-arabinopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Sugar Binding Transporter Arad_9553 from Agrobacterium Radiobacter, Target Efi-511541
To be Published
|
|
4PPE
 
 | | human RNF4 RING domain | | Descriptor: | E3 ubiquitin-protein ligase RNF4, ZINC ION | | Authors: | Perry, J.J, Arvai, A.S, Hitomi, C, Tainer, J.A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response.
Embo Rep., 15, 2014
|
|
4PRF
 
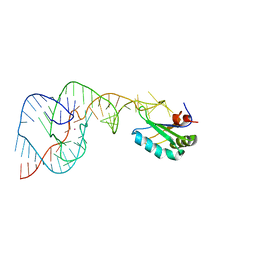 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PRU
 
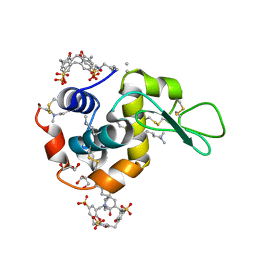 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Lysozyme C | | Authors: | McGovern, R.E, Lyons, J.A, Crowley, P.B. | | Deposit date: | 2014-03-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
7MD7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with triphenylphosphonium analog of chloramphenicol CAM-C4-TPP and protein Y (YfiA) at 2.80A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Pavlova, J.A, Lukianov, D.A, Tereshchenkov, A.G, Makarov, G.I, Khairullina, Z.Z, Tashlitsky, V.N, Paleskava, A, Konevega, A.L, Bogdanov, A.A, Osterman, I.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2021-04-03 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.
Antibiotics, 10, 2021
|
|
4PTS
 
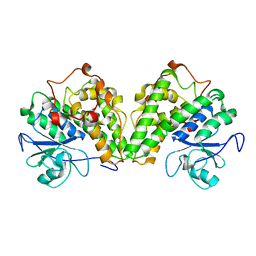 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | Descriptor: | glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
3QKS
 
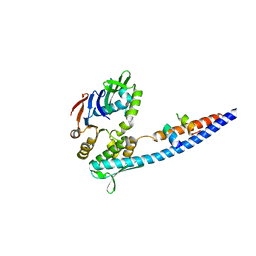 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4PFB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Fusobacterium nucleatun (FN1258, TARGET EFI-510120) with bound SN-glycerol-3-phosphate | | Descriptor: | C4-dicarboxylate-binding protein, IMIDAZOLE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3QKT
 
 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4PF6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS (RD1_0742, TARGET EFI-510239) WITH BOUND 3-DEOXY-D-MANNO-OCT-2-ULOSONIC ACID (KDO) | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, C4-dicarboxylate-binding protein, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PGN
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND INDOLE PYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3QB3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92KL25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sue, G, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Relocation of internal ionizable residues to cavities created by single alanine substitutions
To be Published
|
|
4PED
 
 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
3Q7Q
 
 | | Crystal Structure of Rad G-domain Q148A-GTP Analog Complex | | Descriptor: | CALCIUM ION, GTP-binding protein RAD, MAGNESIUM ION, ... | | Authors: | Sasson, Y, Navon-Perry, L, Hirsch, J.A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
4PEL
 
 | | S1C mutant of Penicillin G acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase subunit alpha, Penicillin G acylase subunit beta | | Authors: | Ramasamy, S, Chand, D, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin G acylase
To Be Published
|
|
