6UPS
 
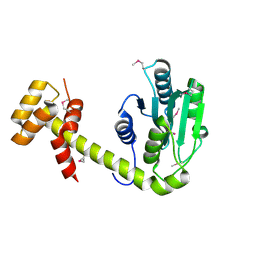 | |
3L40
 
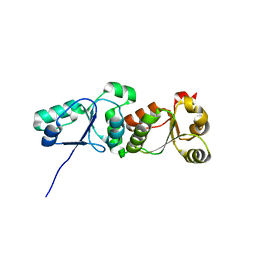 | |
5VFC
 
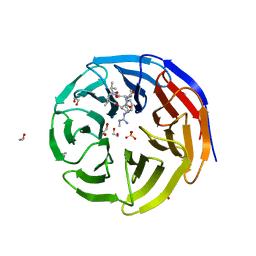 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
6UPU
 
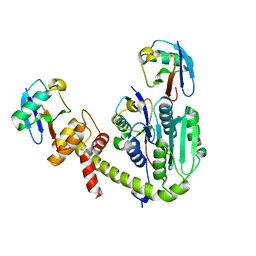 | |
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
3LQ6
 
 | |
3D03
 
 | | 1.9A structure of Glycerophoshphodiesterase (GpdQ) from Enterobacter aerogenes | | Descriptor: | COBALT (II) ION, Phosphohydrolase | | Authors: | Hadler, K.S, Tanifum, E, Yip, S.H.-C, Miti, N, Guddat, L.W, Jackson, C.J, Gahan, L.R, Carr, P.D, Nguyen, K, Ollis, D.L, Hengge, A.C, Larrabee, J.A, Schenk, G. | | Deposit date: | 2008-04-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-promoted formation of a catalytically competent binuclear center and regulation of reactivity in a glycerophosphodiesterase from Enterobacter aerogenes.
J.Am.Chem.Soc., 130, 2008
|
|
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5KCD
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
3CT7
 
 | | Crystal structure of D-allulose 6-phosphate 3-epimerase from Escherichia Coli K-12 | | Descriptor: | D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
5VMQ
 
 | |
5VGS
 
 | | Crystal structure of lachrymatory factor synthase from Allium cepa in complex with crotyl alcohol | | Descriptor: | (2E)-but-2-en-1-ol, (2Z)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Silvaroli, J.A, Pleshinger, M.J, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzyme That Makes You Cry-Crystal Structure of Lachrymatory Factor Synthase from Allium cepa.
ACS Chem. Biol., 12, 2017
|
|
3CO3
 
 | | X-Ray Crystal Structure of a Monofunctional Platinum-DNA Adduct, cis-{Pt(NH3)2(pyridine)}2+ Bound to Deoxyguanosine in a Dodecamer Duplex | | Descriptor: | 5'-D(*DCP*DCP*DTP*DCP*DTP*DCP*DGP*DTP*DCP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DGP*DAP*DGP*DAP*DGP*DG)-3', cis-diammine(pyridine)chloroplatinum(II) | | Authors: | Lovejoy, K.S, Todd, R.C, Zhang, S, McCormick, M.S, D'Aquino, J.A, Reardon, J.T, Sancar, A, Giacomini, K.M, Lippard, S.J. | | Deposit date: | 2008-03-27 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5KCF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
3CSU
 
 | | CATALYTIC TRIMER OF ESCHERICHIA COLI ASPARTATE TRANSCARBAMOYLASE | | Descriptor: | CALCIUM ION, PROTEIN (ASPARTATE CARBAMOYLTRANSFERASE) | | Authors: | Beernink, P.T, Endrizzi, J.A, Alber, T, Schachman, H.K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3LI0
 
 | | Crystal structure of the mutant R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3CRW
 
 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
5VZL
 
 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
3LI1
 
 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLD
 
 | | Crystal structure of the mutant S127G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
5W3V
 
 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
3LHT
 
 | | Crystal structure of the mutant V201F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
