8RGW
 
 | | Serial synchrotron in plate room temperature structure of Dye Type Peroxidase Aa, 12 drops merged | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thompson, A.J, Hough, M.A, Williams, L.J, Worrall, J.A.R, Sanchez-Weatherby, J, Mikolajek, H, Sandy, J. | | Deposit date: | 2023-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2K0C
 
 | | Zinc-finger 2 of Nup153 | | Descriptor: | Nuclear pore complex protein Nup153, ZINC ION | | Authors: | Bangert, J.A, Schrader, N, Vetter, I.R, Stoll, R. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex.
Structure, 16, 2008
|
|
2JV9
 
 | |
8F9Y
 
 | | SAL1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SAL1 phosphatase | | Authors: | Frkic, R.L, Kaczmarski, J.A, Tan, L, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2JVI
 
 | |
3QS2
 
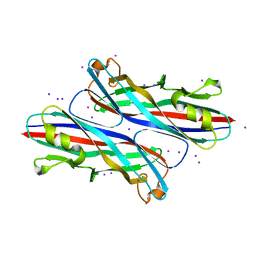 | |
6VPS
 
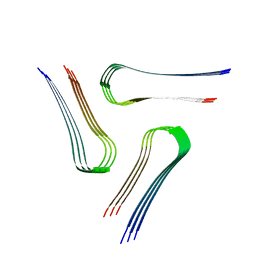 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
6SI5
 
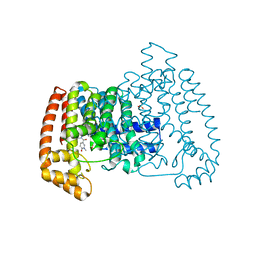 | | T. cruzi FPPS in complex with 1-methyl-5-(4,5,6,7-tetrahydrothieno[3,2-c]pyridine-5-carbonyl)pyridin-2(1H)-one | | Descriptor: | 5-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylcarbonyl)-1-methyl-pyridin-2-one, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Cornaciu, I, Clavel, D, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
2L68
 
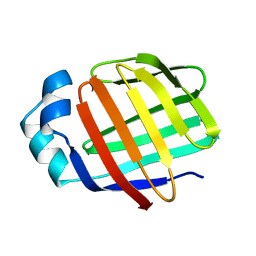 | | Solution Structure of Human Holo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L96
 
 | | Solution structure of LAK160-P7 | | Descriptor: | LAK160-P7 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
6TAS
 
 | | Structure of the two-fold capsomer of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
2KZZ
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
1ISC
 
 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, FE (III) ION, IRON(III) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
2KZM
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2MZC
 
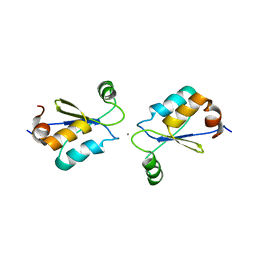 | | Metal Binding of Glutaredoxins | | Descriptor: | Glutaredoxin, SILVER ION | | Authors: | Bilinovich, S.M, Caporoso, J.A, Taraboletti, A, Duangjumpa, N, Panzner, M.J, Prokop, J.W, Shriver, L.P, Leeper, T.C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal Binding of Glutaredoxins
To be Published
|
|
1TTC
 
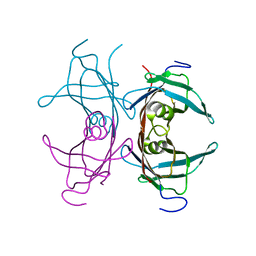 | |
3RCZ
 
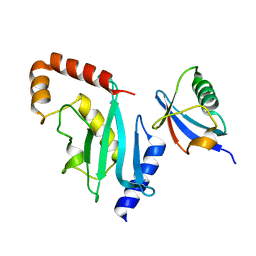 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
2NO2
 
 | |
2N5C
 
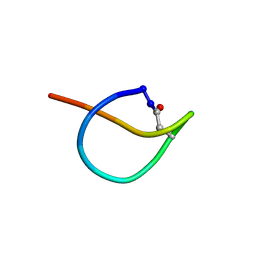 | | Solution NMR structure of the lasso peptide chaxapeptin | | Descriptor: | chaxapeptin | | Authors: | Elsayed, S.S, Trusch, F, Deng, H, Raab, A, Prokes, I, Busarakam, K, Asenjo, J.A, Andrews, B.A, van West, P, Bull, A.T, Goodfellow, M, Yi, Y, Ebel, R, Jaspars, M, Rateb, M.E. | | Deposit date: | 2015-07-14 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert.
J.Org.Chem., 80, 2015
|
|
2K57
 
 | | Solution NMR Structure of Putative Lipoprotein from Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A. | | Descriptor: | Putative Lipoprotein | | Authors: | Hang, D, Aramini, J.A, Rossi, P, Wang, D, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-25 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Putative Lipoprotein from
Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A.
To be Published
|
|
2KAE
 
 | | data-driven model of MED1:DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | Authors: | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
1GID
 
 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
2NNA
 
 | | Structure of the MHC class II molecule HLA-DQ8 bound with a deamidated gluten peptide | | Descriptor: | MHC class II antigen, gluten peptide | | Authors: | Henderson, K.N, Tye-Din, J.A, Rossjohn, J, Anderson, R.P. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease
Immunity, 27, 2007
|
|
2N4X
 
 | |
1XA8
 
 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
