1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2NMS
 
 | |
2YOG
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- alpha-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxol an-2-yl]methyl]thiourea, THYMIDYLATE KINASE | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
2NQH
 
 | |
2HXZ
 
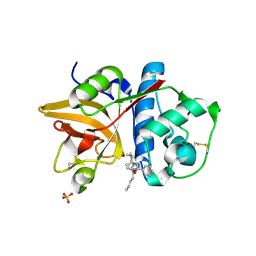 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
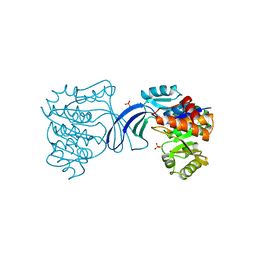
1RKS
 
 | |
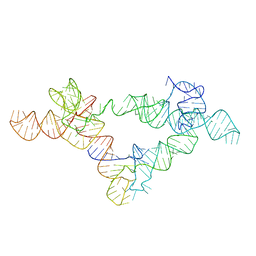
6UFH
 
 | |
6SI5
 
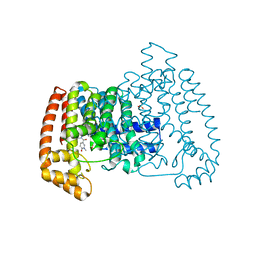 | | T. cruzi FPPS in complex with 1-methyl-5-(4,5,6,7-tetrahydrothieno[3,2-c]pyridine-5-carbonyl)pyridin-2(1H)-one | | Descriptor: | 5-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylcarbonyl)-1-methyl-pyridin-2-one, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Cornaciu, I, Clavel, D, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
2K0C
 
 | | Zinc-finger 2 of Nup153 | | Descriptor: | Nuclear pore complex protein Nup153, ZINC ION | | Authors: | Bangert, J.A, Schrader, N, Vetter, I.R, Stoll, R. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex.
Structure, 16, 2008
|
|
2W1H
 
 | | Fragment-Based Discovery of the Pyrazol-4-yl urea (AT9283), a Multi- targeted Kinase Inhibitor with Potent Aurora Kinase Activity | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[3-(1H-BENZIMIDAZOL-2-YL)-1H-PYRAZOL-4-YL]BENZAMIDE | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2JVJ
 
 | |
2VTA
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 1H-indazole, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
1XU1
 
 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1JQG
 
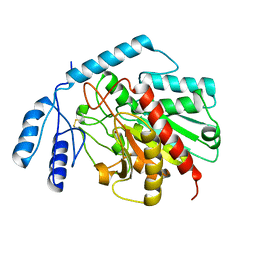 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
2K3S
 
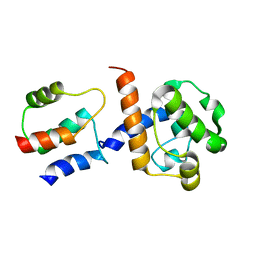 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
2FLI
 
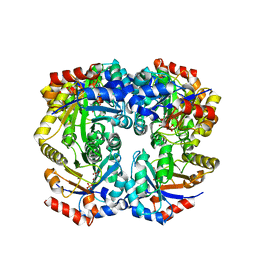 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | Descriptor: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
1JGL
 
 | |
2JVI
 
 | |
2WWD
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2JV9
 
 | |
2FFL
 
 | |
2FLJ
 
 | | Fatty acid binding protein from locust flight muscle in complex with oleate | | Descriptor: | Fatty acid-binding protein, OLEIC ACID | | Authors: | Luecke, C, Qiao, Y, van Moerkerk, H.T.B, Veerkamp, J.H, Hamilton, J.A. | | Deposit date: | 2006-01-06 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fatty-acid-binding protein from the flight muscle of Locusta migratoria: evolutionary variations in fatty acid binding.
Biochemistry, 45, 2006
|
|
368D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L, Malinina, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1Y7M
 
 | | Crystal Structure of the B. subtilis YkuD protein at 2 A resolution | | Descriptor: | CADMIUM ION, SULFATE ION, hypothetical protein BSU14040 | | Authors: | Bielnicki, J.A, Devedjiev, Y, Derewenda, U, Dauter, Z, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes.
Proteins, 62, 2006
|
|
