4IJI
 
 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
5Q6B
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 148) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4IHQ
 
 | | Archaellum Assembly ATPase FlaI bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, FlaI ATPase, ... | | Authors: | Reindl, S, Williams, G.J, Tainer, J.A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics.
Mol.Cell, 49, 2013
|
|
3P2R
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate | | Descriptor: | Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
4ID9
 
 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1 | | Descriptor: | ALANINE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1
To be Published
|
|
4IP4
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg | | Descriptor: | CARBON DIOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with mg
To be Published
|
|
3PDX
 
 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
3PBV
 
 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
4IT1
 
 | | Crystal structure of enolase pfl01_3283 (target efi-502286) from pseudomonas fluorescens pf0-1 with bound magnesium, potassium and tartrate | | Descriptor: | BICARBONATE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Enolase Pfl01_3283 from Pseudomonas Fluorescens
To be Published
|
|
4ISD
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE homolog from BURKHOLDERIA GL BGR1, TARGET EFI-501803, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of GLUTATHIONE TRANSFERASE homolog from BURKHOLDERIA GL BGR1, TARGET EFI-501803, with bound glutathione
TO BE PUBLISHED
|
|
1PTZ
 
 | | Crystal structure of the human CU, Zn Superoxide Dismutase, Familial Amyotrophic Lateral Sclerosis (FALS) Mutant H43R | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
4IV2
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY-derivative, 5a | | Descriptor: | 4-[1-(2-methylpropyl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IVF
 
 | | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit | | Descriptor: | CITRIC ACID, GLUTATHIONE, Putative uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit
To be Published
|
|
4IW8
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY-derivative, 9a | | Descriptor: | 4-[1-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0375 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4J2F
 
 | | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866 | | Descriptor: | GLYCEROL, Glutathione s-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-02-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
TO BE PUBLISHED
|
|
3PFR
 
 | | Crystal structure of D-Glucarate dehydratase related protein from Actinobacillus Succinogenes complexed with D-Glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Mills-Groninger, F, Ghasempur, S, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase related protein from Actinobacillus Succinogenes complexed with D-Glucarate
To be Published
|
|
4IUI
 
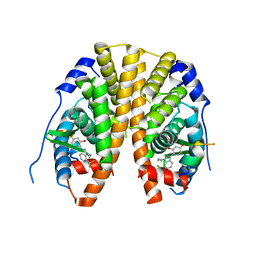 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY derivative, 4a | | Descriptor: | 4-[1-butyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
1RQ2
 
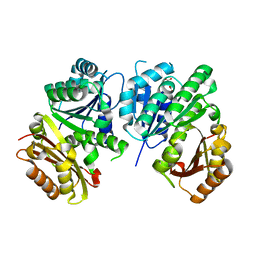 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
4IY0
 
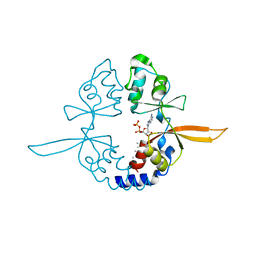 | | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4 | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Encinar, J.A, Spiwok, V, Gomez-Garcia, I, Oyenarte, I, Ereno-Orbea, J, Terashima, H, Accardi, A, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To be Published
|
|
1RTX
 
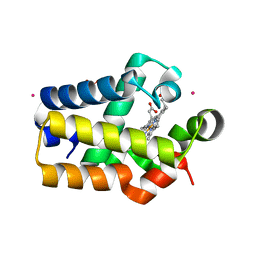 | | Crystal Structure of Synechocystis Hemoglobin with a Covalent Heme Linkage | | Descriptor: | CADMIUM ION, Cyanoglobin, POTASSIUM ION, ... | | Authors: | Hoy, J.A, Kundu, S, Trent, J.T, Ramaswamy, S, Hargrove, M.S. | | Deposit date: | 2003-12-10 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of synechocystis hemoglobin with a covalent heme linkage.
J.Biol.Chem., 279, 2004
|
|
4J1O
 
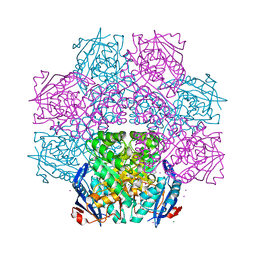 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
1RJ1
 
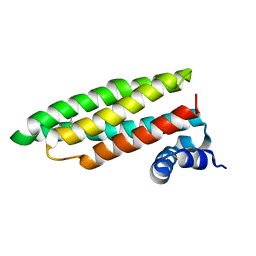 | | Crystal Structure of a Cell Wall Invertase Inhibitor from Tobacco | | Descriptor: | invertase inhibitor | | Authors: | Hothorn, M, D'Angelo, I, Marquez, J.A, Greiner, S, Scheffzek, K. | | Deposit date: | 2003-11-18 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension
J.Mol.Biol., 335, 2004
|
|
3PQZ
 
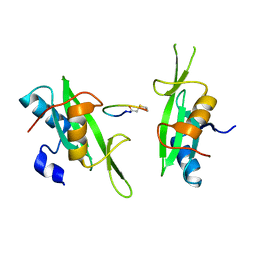 | | Grb7 SH2 with peptide | | Descriptor: | Growth factor receptor-bound protein 7, cyclic peptide | | Authors: | Wilce, J.A. | | Deposit date: | 2010-11-29 | | Release date: | 2011-07-20 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis of binding by cyclic nonphosphorylated Peptide antagonists of grb7 implicated in breast cancer progression
J.Mol.Biol., 412, 2011
|
|
3PSX
 
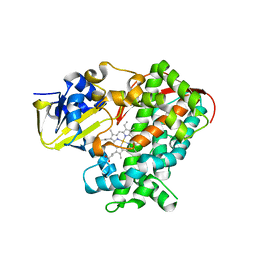 | | Crystal structure of the KT2 mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Yorke, J.A, Bell, S.G, Zhou, W, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure, electronic properties and catalytic behaviour of an activity-enhancing CYP102A1 (P450(BM3)) variant
Dalton Trans, 40, 2011
|
|
1RLU
 
 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein ftsZ, GLYCEROL | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-11-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
