2YNX
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Archaea Common Ancestor (LACA) from the Precambrian Period | | Descriptor: | ACETATE ION, LACA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
2H75
 
 | |
2GVG
 
 | | Crystal Structure of human NMPRTase and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HBQ
 
 | |
399D
 
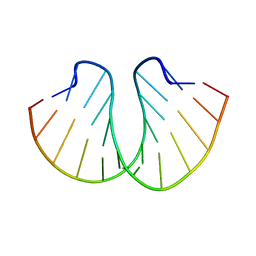 | | STRUCTURE OF D(CGCCCGCGGGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*G)-3') | | Authors: | Malinina, L, Fernandez, L.G, Subirana, J.A. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the d(CGCCCGCGGGCG) dodecamer: a kinked A-DNA molecule showing some B-DNA features.
J.Mol.Biol., 285, 1999
|
|
2OIH
 
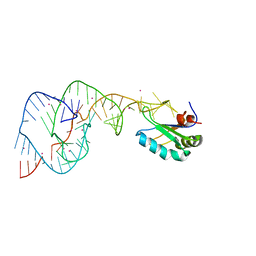 | | Hepatitis Delta Virus gemonic ribozyme precursor with C75U mutation and bound to monovalent cation Tl+ | | Descriptor: | HDV ribozyme, THALLIUM (I) ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
1FKL
 
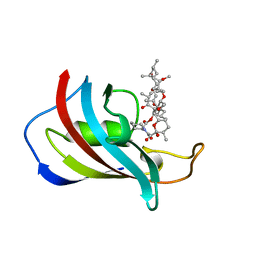 | | ATOMIC STRUCTURE OF FKBP12-RAPAYMYCIN, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKJ
 
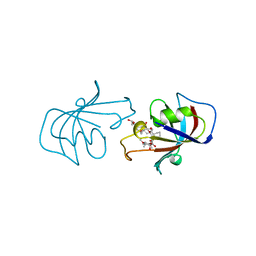 | | ATOMIC STRUCTURE OF FKBP12-FK506, AN IMMUNOPHILIN IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
370D
 
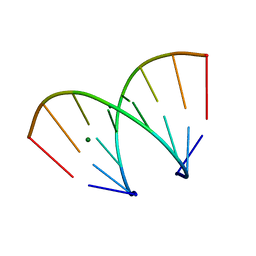 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
392D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
2I7B
 
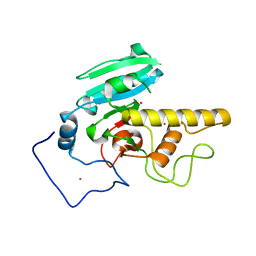 | |
371D
 
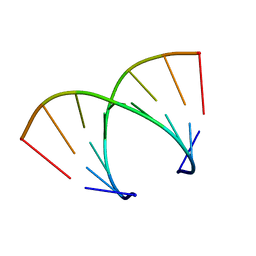 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1V0A
 
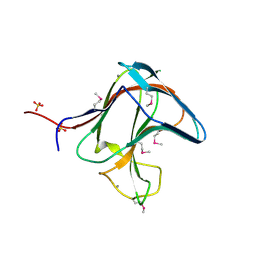 | | Family 11 Carbohydrate-Binding Module of cellulosomal cellulase Lic26A-Cel5E of Clostridium thermocellum | | Descriptor: | CALCIUM ION, ENDOGLUCANASE H, SULFATE ION | | Authors: | Carvalho, A.L, Romao, M.J, Goyal, A, Prates, J.A.M, Pires, V.M.R, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accomodates Beta-1,4- and Beta-1,3-1,4-Mixed Linked Glucans at a Single Binding Site
J.Biol.Chem., 279, 2004
|
|
1G5H
 
 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
2YN4
 
 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YNE
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor | | Descriptor: | 2-(3-piperidin-4-yloxy-1-benzothiophen-2-yl)-5-[(1,3,5-trimethylpyrazol-4-yl)methyl]-1,3,4-oxadiazole, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2Y8Y
 
 | |
2Y8W
 
 | |
2Y9H
 
 | |
2YEV
 
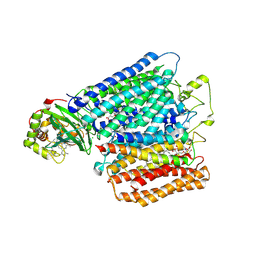 | | Structure of caa3-type cytochrome oxidase | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, (2R)-3-HYDROXYPROPANE-1,2-DIYL DIHEXADECANOATE, ... | | Authors: | Lyons, J.A, Aragao, D, Soulimane, T, Caffrey, M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Electron Transfer in Caa3-Type Cytochrome Oxidases.
Nature, 487, 2012
|
|
1G39
 
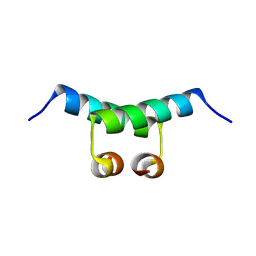 | | WILD-TYPE HNF-1ALPHA DIMERIZATION DOMAIN | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G5I
 
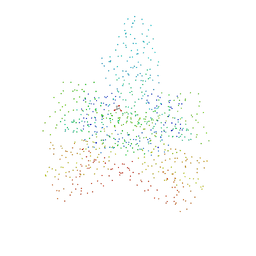 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
1UY0
 
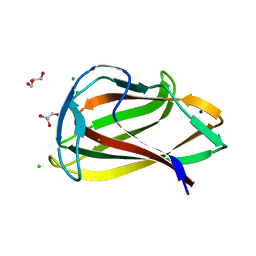 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
2KZM
 
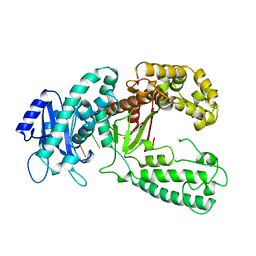 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2OJ3
 
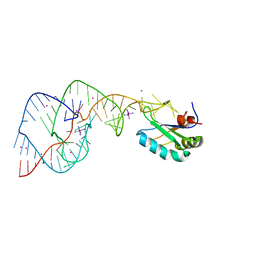 | | Hepatitis Delta Virus ribozyme precursor structure, with C75U mutation, bound to Tl+ and cobalt hexammine (Co(NH3)63+) | | Descriptor: | COBALT HEXAMMINE(III), HDV RIBOZYME, THALLIUM (I) ION, ... | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
