4MNC
 
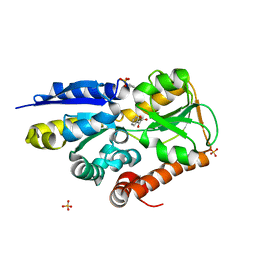 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | 分子名称: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-09-10 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MPI
 
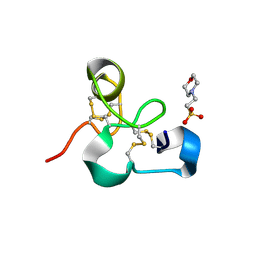 | | Crystal structure of the chitin-binding module (CBM18) of a chitinase-like protein from Hevea brasiliensis | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Class I chitinase | | 著者 | Martinez-Caballero, C.S, Hermoso, J.A, Rodriguez-Romero, A. | | 登録日 | 2013-09-12 | | 公開日 | 2014-08-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Comparative study of two GH19 chitinase-like proteins from Hevea brasiliensis, one exhibiting a novel carbohydrate-binding domain.
Febs J., 281, 2014
|
|
4QOJ
 
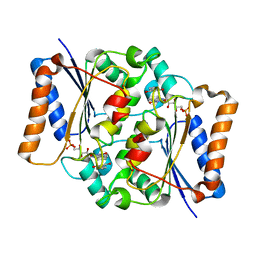 | | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A | | 分子名称: | FLAVIN MONONUCLEOTIDE, RESVERATROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A
To be Published
|
|
4QP5
 
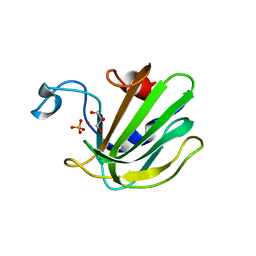 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate | | 分子名称: | GLYCEROL, Lysostaphin, PHOSPHATE ION, ... | | 著者 | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | 登録日 | 2014-06-22 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4Q60
 
 | | Crystal structure of a 4-hydroxyproline epimerase from Burkholderia Multivorans atcc 17616, target EFI-506586, open form, with bound pyrrole-2-carboxylate | | 分子名称: | GLYCEROL, PROLINE RACEMASE, PYRROLE-2-CARBOXYLATE | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-04-18 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF PROLINE RACEMASE Bmul_4447 FROM Burkholderia multivorans, TARGET EFI-506586
To be Published
|
|
4QGK
 
 | |
4QOD
 
 | | The value crystal structure of apo quinone reductase 2 at 1.35A | | 分子名称: | GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The value crystal structure of apo quinone reductase 2 at 1.35A
To be Published
|
|
4QOI
 
 | | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A | | 分子名称: | FLAVIN MONONUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A
To be Published
|
|
4QPB
 
 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | 分子名称: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | 著者 | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | 登録日 | 2014-06-22 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4QOG
 
 | | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A
To be Published
|
|
4QOE
 
 | | The value 'crystal structure of fad quinone reductase 2 at 1.45A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The value 'crystal structure of fad quinone reductase 2 at 1.45A
To be Published
|
|
3E4R
 
 | | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nitrate transport protein | | 著者 | Balan, A, Araujo, F.T, Sanches, M, Chirgadze, D.Y, Blundell, T.B, Barbosa, J.A.R.G. | | 登録日 | 2008-08-12 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES
To be Published
|
|
4Q3W
 
 | | Crystal structure of C. violaceum phenylalanine hydroxylase D139E mutation | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, Phenylalanine-4-hydroxylase | | 著者 | Ronau, J.A, Abu-Omar, M.M, Das, C. | | 登録日 | 2014-04-12 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A conserved acidic residue in phenylalanine hydroxylase contributes to cofactor affinity and catalysis.
Biochemistry, 53, 2014
|
|
4Q3X
 
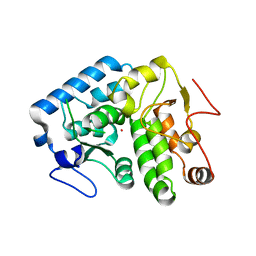 | |
4Q37
 
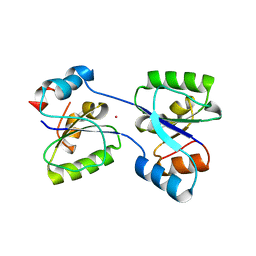 | |
4Q9R
 
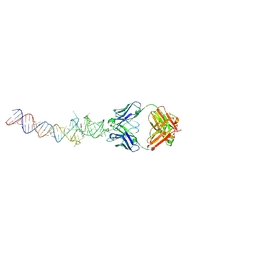 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | 著者 | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | 登録日 | 2014-05-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4R3R
 
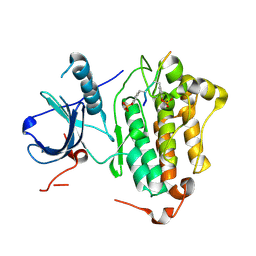 | | Crystal structures of EGFR in complex with Mig6 | | 分子名称: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | 著者 | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | 登録日 | 2014-08-17 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RBM
 
 | | Porphyromonas gingivalis gingipain K (Kgp) catalytic and immunoglobulin superfamily-like domains | | 分子名称: | (3S)-3,7-diaminoheptan-2-one, ACETATE ION, AZIDE ION, ... | | 著者 | de Diego, I, Veillard, F, Sztukowska, M.N, Guevara, T, Potempa, B, Pomowski, A, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | 登録日 | 2014-09-12 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and Mechanism of Cysteine Peptidase Gingipain K (Kgp), a Major Virulence Factor of Porphyromonas gingivalis in Periodontitis.
J.Biol.Chem., 289, 2014
|
|
4O94
 
 | | Crystal structure of a trap periplasmic solute binding protein from Rhodopseudomonas palustris HaA2 (RPB_3329), Target EFI-510223, with bound succinate | | 分子名称: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter DctP subunit | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-01-01 | | 公開日 | 2014-01-22 | | 最終更新日 | 2015-02-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | 分子名称: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-11 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | 分子名称: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-11 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | 分子名称: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-28 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-25 | | 公開日 | 2014-03-05 | | 最終更新日 | 2015-02-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4R2F
 
 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | 分子名称: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-11 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
4R6Y
 
 | | Crystal structure of solute-binding protein stm0429 from salmonella enterica subsp. enterica serovar typhimurium str. lt2, target efi-510776, a closed conformation, in complex with glycerol and acetate | | 分子名称: | ACETATE ION, GLYCEROL, Putative 2-aminoethylphosphonate-binding periplasmic protein | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-26 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Crystal Structure of Transporter STM0429 from Salmonella Enterica
To be Published
|
|
