3OS8
 
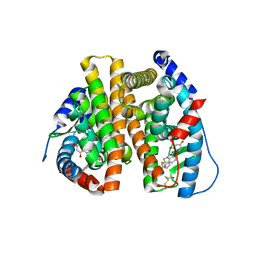 | | Estrogen Receptor | | Descriptor: | 4-[1-benzyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
4IGJ
 
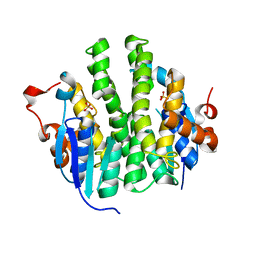 | | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175 | | Descriptor: | Maleylacetoacetate isomerase, SULFATE ION | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175
To be Published
|
|
4IHC
 
 | | Crystal structure of probable mannonate dehydratase Dd703_0947 (target EFI-502222) from Dickeya dadantii Ech703 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mannonate dehydratase Dd703_0947 from Dickeya dadantii Ech703
To be Published
|
|
4IBO
 
 | | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446) | | Descriptor: | CHLORIDE ION, Gluconate dehydrogenase, MAGNESIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446)
To be Published
|
|
3OVM
 
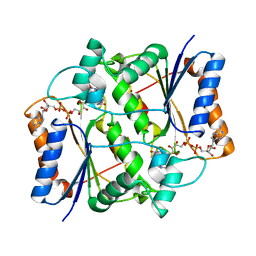 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
1QH0
 
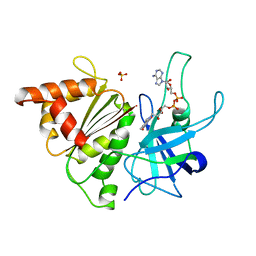 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
3P60
 
 | | Crystal structure of the mutant T159V of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
5Q1R
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000291a | | Descriptor: | 3-(azepan-1-ylmethyl)-1~{H}-indole, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q28
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000186a | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, NICKEL (II) ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2R
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 19) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1QM4
 
 | | Methionine Adenosyltransferase Complexed with a L-Methionine Analogue | | Descriptor: | L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, METHIONINE ADENOSYLTRANSFERASE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Tetrameric Methionine Adenosyltransferase from Rat Liver Reveals the Methionine-Binding Site
J.Mol.Biol., 300, 2000
|
|
5Q38
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 36) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3H
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 45) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3OTU
 
 | | PDK1 mutant bound to allosteric disulfide fragment activator JS30 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(naphthalen-1-ylmethyl)piperazin-1-yl]-4-oxobutane-1-thiol, ... | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1013 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1Q8Z
 
 | | The apoenzyme structure of the yeast SR protein kinase, Sky1p | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, SR Protein Kinase, ... | | Authors: | Nolen, B, Ngo, J, Chakrabarti, S, Vu, D, Adams, J.A, Ghosh, G. | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Nucleotide-Induced Conformational Changes in the Saccharomyces cerevisiae SR Protein Kinase, Sky1p, Revealed by X-Ray Crystallography
Biochemistry, 42, 2003
|
|
5Q20
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000524a | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1QTW
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
5Q2G
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 8) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2W
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 24) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3OW1
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with MG | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | CRYSTAL STRUCTURE OF D-MANNONATE DEHYDRATASE FROM CHROMOHALOBACTER SALEXIGENS complexed with MG
To be Published
|
|
5Q3C
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 40) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3S
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 56) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q48
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 73) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q4O
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 89) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q54
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 105) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
