5F12
 
 | | WrbA in complex with FMN under crystallization conditions of WrbA-FMN-BQ structure (4YQE) | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Kuta Smatanova, I, Carey, J, Ettrich, R. | | Deposit date: | 2015-11-29 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
6F23
 
 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
8Z1L
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
4CEW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5EWP
 
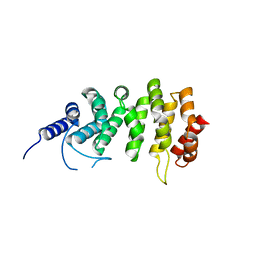 | | ARO (armadillo repeats only protein) from Plasmodium falciparum | | Descriptor: | ARO (armadillo repeats only protein) | | Authors: | Brown, C, Zhang, K, Emery, J, Prusty, D, Wetzel, J, Heincke, D, Gilberger, T, Junop, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ARO (armadillo repeats only protein) from Plasmodium falciparum
To Be Published
|
|
7B85
 
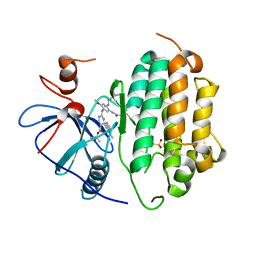 | | Crystal Structure of EGFR-WT in Complex with TAK-788 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-12-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
4G43
 
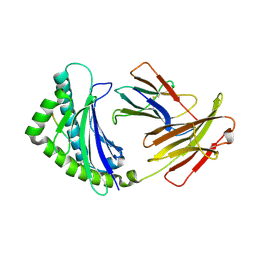 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
5FBH
 
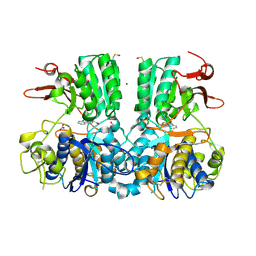 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
4Q9C
 
 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4H0N
 
 | | Crystal structure of Spodoptera frugiperda DNMT2 E260A/E261A/K263A mutant | | Descriptor: | CALCIUM ION, DNMT2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, S, Du, J, Yang, H, Yin, J, Zhong, J, Ding, J. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Functional and structural characterization of DNMT2 from Spodoptera frugiperda.
J Mol Cell Biol, 5, 2013
|
|
4LVG
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-N-[4-(phenylsulfonyl)phenyl]-2-(pyridin-3-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M6Q
 
 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M6P
 
 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | N-[4-(phenylsulfonyl)benzyl]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3BE4
 
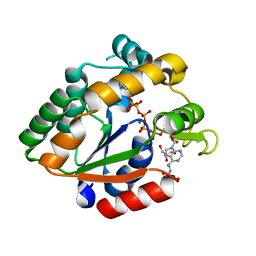 | | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, GLYCEROL, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360.
To be Published
|
|
7YTJ
 
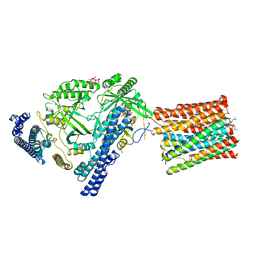 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
3N9X
 
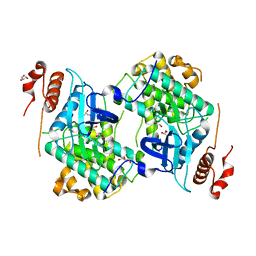 | | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0 | | Descriptor: | GLYCEROL, Phosphotransferase | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Artz, J, Amani, M, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0
To be Published
|
|
4Q9B
 
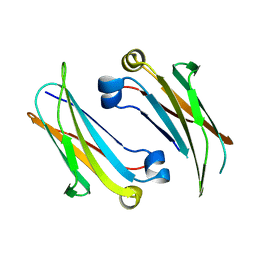 | | IgNAR antibody domain C2 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WZM
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
4LVD
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-nitrophenyl)cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4Q97
 
 | | IgNAR antibody domain C1 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4LVB
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(acetylamino)phenyl]cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3B6N
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax.
To be Published
|
|
3TTI
 
 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
