1MMX
 
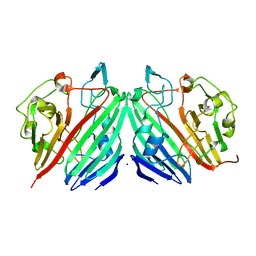 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1MQA
 
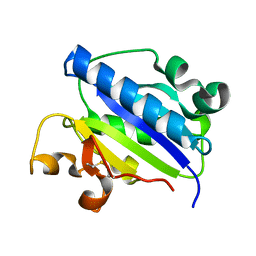 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MOM
 
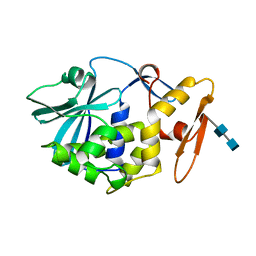 | | CRYSTAL STRUCTURE OF MOMORDIN, A TYPE I RIBOSOME INACTIVATING PROTEIN FROM THE SEEDS OF MOMORDICA CHARANTIA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOMORDIN | | Authors: | Husain, J, Tickle, I.J, Wood, S.P. | | Deposit date: | 1994-03-04 | | Release date: | 1994-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia.
FEBS Lett., 342, 1994
|
|
1MP6
 
 | |
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MPG
 
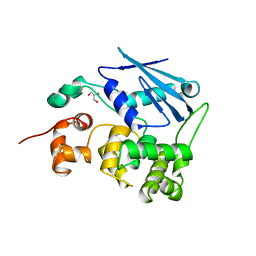 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
6QUR
 
 | |
6QUT
 
 | |
6QW2
 
 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
1MDI
 
 | |
4TXH
 
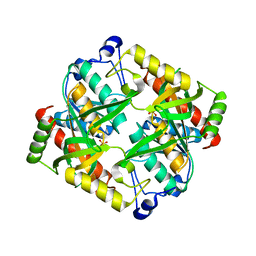 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Torini, J, Romanello, L, Marinho, A, Brandao-Neto, J, Cassago, A, DeMarco, R, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
1MDK
 
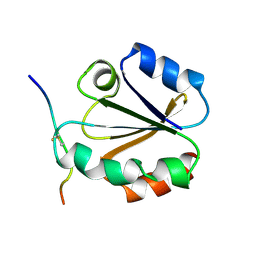 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
1MDJ
 
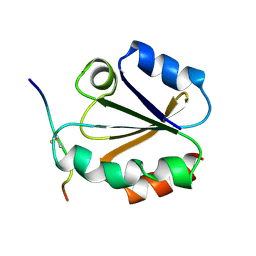 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
6QN2
 
 | |
6QNG
 
 | |
6QW5
 
 | | Structure and function of the toscana virus cap snatching endonuclease | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, RNA-dependent RNA polymerase, ... | | Authors: | Reguera, J, Jones, R, Bragagniolo, G, Lessoued, S, Mate, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-25 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structure and function of the Toscana virus cap-snatching endonuclease.
Nucleic Acids Res., 47, 2019
|
|
4TXJ
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with thymidine | | Descriptor: | SULFATE ION, THYMIDINE, Uridine phosphorylase | | Authors: | Torini, J, Marinho, A, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
6QX7
 
 | |
6QXM
 
 | | Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6QN0
 
 | |
1MTR
 
 | | HIV-1 PROTEASE COMPLEXED WITH A CYCLIC PHE-ILE-VAL PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Wickramasinghe, W, Begun, J, Martin, J.L. | | Deposit date: | 1996-02-15 | | Release date: | 1996-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-based cyclic peptidomimetics of Phe-Ile-Val that inhibit HIV-1 protease using a novel enzyme-binding mode.
J.Am.Chem.Soc., 118, 1996
|
|
1MRZ
 
 | | Crystal structure of a flavin binding protein from Thermotoga Maritima, TM379 | | Descriptor: | CITRIC ACID, Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-09-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a flavin-binding protein from Thermotoga Maritima
Proteins, 52, 2003
|
|
1MR8
 
 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1MUG
 
 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | Descriptor: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
