7NEQ
 
 | | Structure of tariquidar-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Kowal, J, Locher, K. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Basis of Drug Recognition by the Multidrug Transporter ABCG2.
J.Mol.Biol., 433, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
1R6Z
 
 | | The Crystal Structure of the Argonaute2 PAZ domain (as a MBP fusion) | | Descriptor: | Chimera of Maltose-binding periplasmic protein and Argonaute 2, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Song, J.J, Liu, J, Tolia, N.H, Schneiderman, J, Smith, S.K, Martienssen, R.A, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2003-10-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Nat.Struct.Biol., 10, 2003
|
|
7NL5
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1R,2R,3R,4S,5R)-4-(hydroxymethyl)cyclohexane-1,2,3,5-tetrol, (1R,6S)-5beta-(Hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2beta,3beta,4alpha-triol, Alpha-1,6-mannanase, ... | | Authors: | Schroeder, S, Offen, W.A, Males, A, Jin, Y, De Boer, C, Enotarpi, J, Marino, L, van der Marel, G.A, Florea, B.I, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
7NJG
 
 | | Xylose isomerase grown inside HARE serial crystallography chip | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6RTL
 
 | | BACTERIOPHAGE SPP1 PROCAPSID-II PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, Brasiles, S, El Sadek, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
7NJI
 
 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2RS3
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-ETHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS5
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RM2
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(6-CHLORO-4-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RR1
 
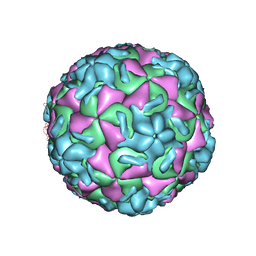 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS1
 
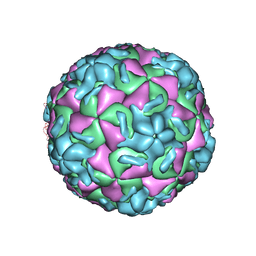 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
3NNG
 
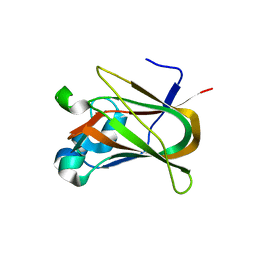 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | Descriptor: | CALCIUM ION, uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
5YLF
 
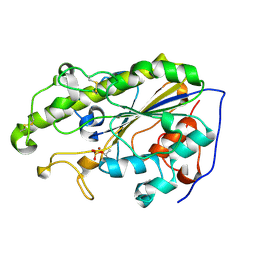 | | MCR-1 complex with D-glucose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
7NLY
 
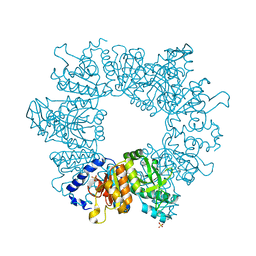 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-Chlorobenzimidazole. | | Descriptor: | 2-chloranyl-1~{H}-benzimidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLS
 
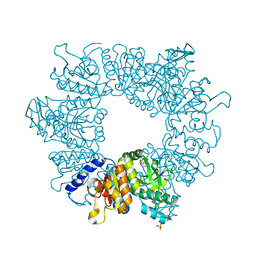 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with methyl 4-hydroxy-3-iodobenzoate | | Descriptor: | Acetylglutamate kinase, SULFATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLT
 
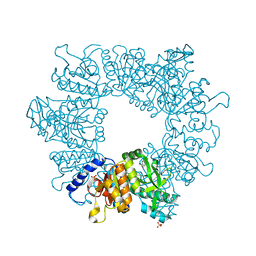 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 4-(4-methylpiperazin-1-yl)benzoic acid | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNB
 
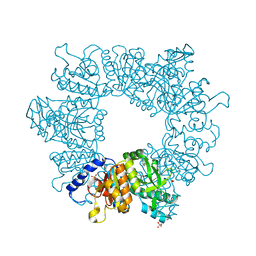 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2,8-bis(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 1,2-ETHANEDIOL, 2,8-bis(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLQ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(isoxazol-5-yl)phenol | | Descriptor: | 2-(1,2-oxazol-5-yl)phenol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-phenyl-1H-imidazole | | Descriptor: | 2-phenyl-1H-imidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN8
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1H-indole-3-carbonitrile. | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indole-3-carbonitrile, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
3CS9
 
 | | Human ABL kinase in complex with nilotinib | | Descriptor: | Nilotinib, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P, Liebetanz, J, Fabbro, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl
Cancer Cell, 7, 2005
|
|
7NLN
 
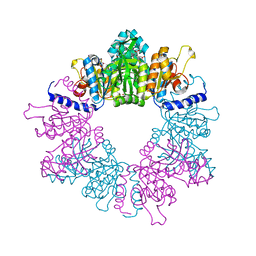 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4DDR
 
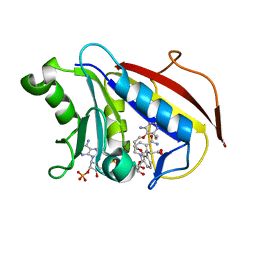 | | Human dihydrofolate reductase complexed with NADPH and P218 | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yuthavong, Y, Tarnchompoo, B, Vilaivan, T, Chitnumsub, P, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Thongphanchang, C, Taweechai, S, Vanichtanankul, J, Rattanajak, R, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7NLO
 
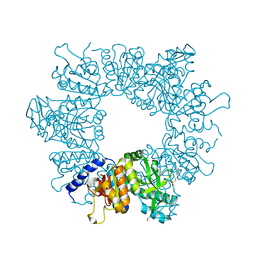 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-arginine | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ARGININE, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
