7SBP
 
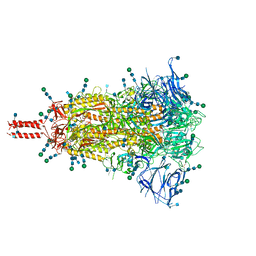 | | Closed state of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBR
 
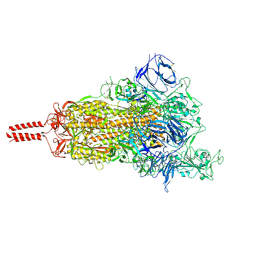 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBO
 
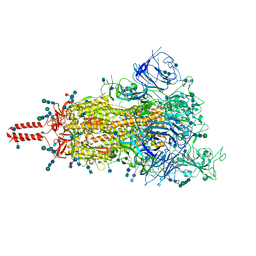 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
8BO6
 
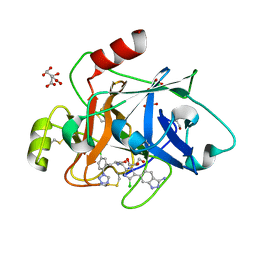 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 2 | | Descriptor: | (~{E})-~{N}-[[5-(3-azanyl-1~{H}-indazol-6-yl)-4-chloranyl-1~{H}-imidazol-2-yl]methyl]-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
7SBT
 
 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
8BO7
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 34 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[[(2~{S})-2-[4-(5-chloranyl-2-cyano-phenyl)-3-methoxy-6-oxidanylidene-2,5-dihydropyridin-1-yl]-3-[(2~{S})-oxan-2-yl]propanoyl]amino]benzoic acid, CITRIC ACID, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
7SBK
 
 | | Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBS
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBL
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7VG3
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a RNA forward strand (5'-phosphorylated), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VG2
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
8BRT
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
6TQU
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
8BRS
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | Descriptor: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
6THC
 
 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP and (4-hydroxyphenyl)(2,3,4-trihydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2,3,4-tris(oxidanyl)phenyl]methanone, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
8BRQ
 
 | | Crystal structure of a surface entropy reduction variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
7SYT
 
 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8BRR
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
4UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6TM0
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
