2FKE
 
 | | FK-506-BINDING PROTEIN: THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX WITH THE ANTAGONIST L-685,818 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Becker, J.W, Mckeever, B.M, Rotonda, J. | | Deposit date: | 1993-01-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818.
J.Biol.Chem., 268, 1993
|
|
3BX4
 
 | | Crystal structure of the snake venom toxin aggretin | | Descriptor: | Aggretin alpha chain, Aggretin beta chain, GLYCEROL, ... | | Authors: | Hooley, E, Papagrigoriou, E, Navdaev, A, Pandey, A, Clemetson, J.M, Clemetson, K.J, Emsley, J. | | Deposit date: | 2008-01-11 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the platelet activator aggretin reveals a novel (alphabeta)2 dimeric structure.
Biochemistry, 47, 2008
|
|
3BX1
 
 | | Complex between the Barley alpha-Amylase/Subtilisin Inhibitor and the subtilisin Savinase | | Descriptor: | Alpha-amylase/subtilisin inhibitor, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Micheelsen, P.O, Vevodova, J, Wilson, K, Skjot, M. | | Deposit date: | 2008-01-11 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mutational Analyses of the Interaction between the Barley alpha-Amylase/Subtilisin Inhibitor and the Subtilisin Savinase Reveal a Novel Mode of Inhibition
J.Mol.Biol., 380, 2008
|
|
2EUY
 
 | | Solution structure of the internal loop of human U65 H/ACA snoRNA 3' hairpin | | Descriptor: | U65 box H/ACA snoRNA | | Authors: | Feigon, J, Khanna, M, Wu, H, Johansson, C, Caizergues-Ferrer, M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural study of the H/ACA snoRNP components Nop10p and the 3' hairpin of U65 snoRNA.
Rna, 12, 2006
|
|
3C1R
 
 | | Crystal structure of oxidized GRX1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutaredoxin-1 | | Authors: | Yu, J, Zhou, C.Z. | | Deposit date: | 2008-01-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glutathionylation-triggered conformational changes of glutaredoxin Grx1 from the yeast Saccharomyces cerevisiae.
Proteins, 72, 2008
|
|
2F0E
 
 | |
2F0I
 
 | |
2F0N
 
 | |
2F0W
 
 | |
3C4A
 
 | | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR158 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable tryptophan hydroxylase vioD | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum.
To be Published
|
|
3C4N
 
 | | Crystal structure of DR_0571 protein from Deinococcus radiodurans in complex with ADP. Northeast Structural Genomics Consortium Target DrR125 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein DR_0571 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Cunningham, K, Owen, L.A, Maglaqui, M, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of DR_0571 protein from Deinococcus radiodurans in complex with ADP.
To be Published
|
|
3C91
 
 | | Thermoplasma acidophilum 20S proteasome with an open gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
2F3R
 
 | |
2F2F
 
 | | Crystal structure of cytolethal distending toxin (CDT) from Actinobacillus actinomycetemcomitans | | Descriptor: | Cytolethal distending toxin A, Cytolethal distending toxin B, cytolethal distending toxin C | | Authors: | Yamada, T, Komoto, J, Saiki, K, Konishi, K, Takusagawa, F. | | Deposit date: | 2005-11-16 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Variation of loop sequence alters stability of cytolethal distending toxin (CDT): crystal structure of CDT from Actinobacillus actinomycetemcomitans
Protein Sci., 15, 2006
|
|
2F3W
 
 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F6U
 
 | |
3BPJ
 
 | | Crystal structure of human translation initiation factor 3, subunit 1 alpha | | Descriptor: | Eukaryotic translation initiation factor 3 subunit J, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Nedyalkova, L, Hong, B, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human translation initiation factor 3, subunit 1 alpha.
To be Published
|
|
2F4X
 
 | | NMR Solution of HIV-1 Lai Kissing Complex | | Descriptor: | 5'-R(P*GP*GP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*C)-3' | | Authors: | Kieken, F, Paquet, F, Brule, F, Paoletti, J, Lancelot, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer
Nucleic Acids Res., 34, 2006
|
|
3BPT
 
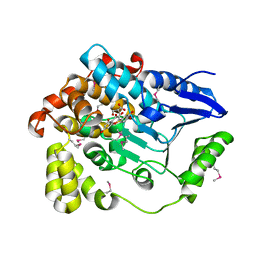 | | Crystal structure of human beta-hydroxyisobutyryl-CoA hydrolase in complex with quercetin | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, 3-hydroxyisobutyryl-CoA hydrolase | | Authors: | Pilka, E.S, Phillips, C, King, O.N.F, Guo, K, von Delft, F, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human beta-hydroxyisobutyryl-CoA hydrolase in complex with quercetin.
To be Published
|
|
3BQ8
 
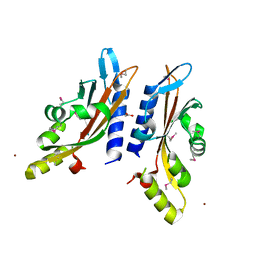 | |
3BMA
 
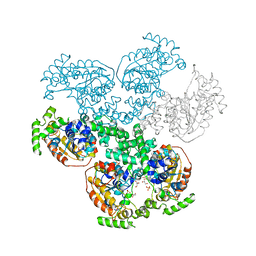 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BNN
 
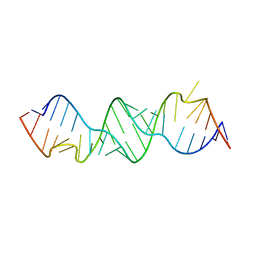 | |
2D1T
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
3BQA
 
 | |
1HH6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
