1LXI
 
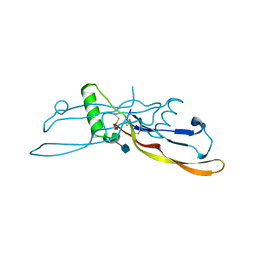 | | Refinement of BMP7 crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BONE MORPHOGENETIC PROTEIN 7 | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-05 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
1LVO
 
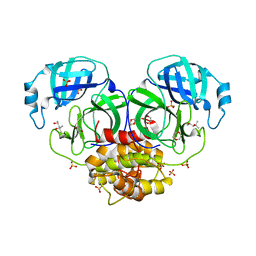 | | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIETHYLENE DIOXIDE, Replicase, ... | | Authors: | Anand, K, Palm, G.J, Mesters, J.R, Siddell, S.G, Ziebuhr, J, Hilgenfeld, R. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain.
EMBO J., 21, 2002
|
|
2OQ1
 
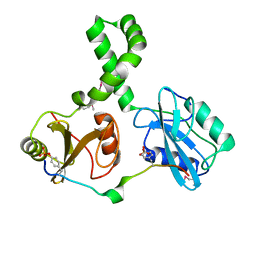 | | Tandem SH2 domains of ZAP-70 with 19-mer zeta1 peptide | | Descriptor: | LEAD (II) ION, T-cell surface glycoprotein CD3 zeta chain, Tyrosine-protein kinase ZAP-70 | | Authors: | Hatada, M.H, Laird, E.R, Green, J, Morgenstern, J, Ram, M.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the interaction of ZAP-70 with the T-cell receptor
Nature, 377, 1995
|
|
1E4I
 
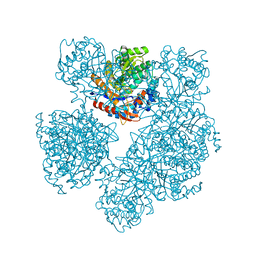 | | 2-deoxy-2-fluoro-beta-D-glucosyl/enzyme intermediate complex of the beta-glucosidase from Bacillus polymyxa | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE | | Authors: | Sanz-Aparicio, J, Gonzalez, B, Hermoso, J.A, Arribas, J.C, Canada, F.J, Polaina, J. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Increased Resistance to Thermal Denaturation Induced by Single Amino Acid Substitution in the Sequence of Beta-Glucosidase a from Bacillus Polymyxa.
Proteins: Struct.,Funct., Genet., 33, 1998
|
|
2OPR
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1WSA
 
 | | STRUCTURE OF L-ASPARAGINASE II PRECURSOR | | Descriptor: | ASPARAGINE AMIDOHYDROLASE | | Authors: | Lubkowski, J, Palm, G.J, Gilliland, G.L, Derst, C, Rohm, K.-H, Wlodawer, A. | | Deposit date: | 1996-08-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and amino acid sequence of Wolinella succinogenes L-asparaginase.
Eur.J.Biochem., 241, 1996
|
|
1WBM
 
 | | HIV-1 protease in complex with symmetric inhibitor, BEA450 | | Descriptor: | (2R,3R,4R,5R)-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]-2,5-BIS(2-PHENYLETHYL)HEXANEDIAMIDE, POL PROTEIN (FRAGMENT) | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease in Complex with Symmetric Inhibitor, Bea450
To be Published
|
|
4G0O
 
 | | Crystal structure of Arabidopsis thaliana AGO5 MID domain | | Descriptor: | Protein argonaute 5, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
1LFO
 
 | | LIVER FATTY ACID BINDING PROTEIN-OLEATE COMPLEX | | Descriptor: | BUTENOIC ACID, LIVER FATTY ACID BINDING PROTEIN, OLEIC ACID, ... | | Authors: | Thompson, J, Winter, N, Terwey, D, Bratt, J, Banaszak, L. | | Deposit date: | 1996-12-09 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates.
J.Biol.Chem., 272, 1997
|
|
4G0Z
 
 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4GI5
 
 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
1XG6
 
 | |
4G0M
 
 | | Crystal structure of Arabidopsis thaliana AGO2 MID domain | | Descriptor: | Protein argonaute 2, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Q
 
 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
2QG1
 
 | | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) | | Descriptor: | 1,2-ETHANEDIOL, Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Salah, E, Phillips, C, Savitsky, P, Boisguerin, P, Oschkinat, H, Gileadi, C, Yang, X, Elkins, J.M, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
To be Published
|
|
1QDM
 
 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
1LX5
 
 | | Crystal Structure of the BMP7/ActRII Extracellular Domain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin Type II Receptor, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-04 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
237L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
1PSR
 
 | | HUMAN PSORIASIN (S100A7) | | Descriptor: | HOLMIUM ATOM, PSORIASIN | | Authors: | Brodersen, D.E, Etzerodt, M, Madsen, P, Celis, J, Thoegersen, H.C, Nyborg, J, Kjeldgaard, M. | | Deposit date: | 1997-11-27 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing.
Structure, 6, 1998
|
|
247L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
1DXL
 
 | | Dihydrolipoamide dehydrogenase of glycine decarboxylase from Pisum Sativum | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Faure, M, Cohen-Addad, C, Bourguignon, J, Macherel, D, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
2A4R
 
 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
