6FW0
 
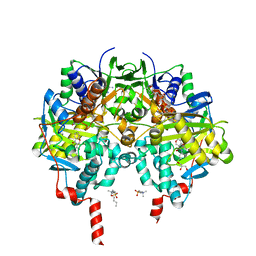 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with chlorophenyl-chromone-carboxamide | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, J, Manzella, N, Cagide, F, Mialet-Perez, J, Uriarte, E, Parini, A, Borges, F, Binda, C. | | Deposit date: | 2018-03-05 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tight-Binding Inhibition of Human Monoamine Oxidase B by Chromone Analogs: A Kinetic, Crystallographic, and Biological Analysis.
J. Med. Chem., 61, 2018
|
|
6FWC
 
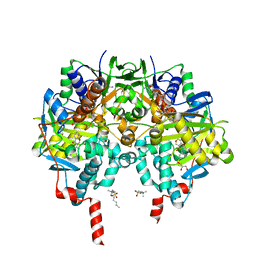 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with fluorophenyl-chromone-carboxamide | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, J, Manzella, N, Cagide, F, Mialet-Perez, J, Uriarte, E, Parini, A, Borges, F, Binda, C. | | Deposit date: | 2018-03-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tight-Binding Inhibition of Human Monoamine Oxidase B by Chromone Analogs: A Kinetic, Crystallographic, and Biological Analysis.
J. Med. Chem., 61, 2018
|
|
6ILZ
 
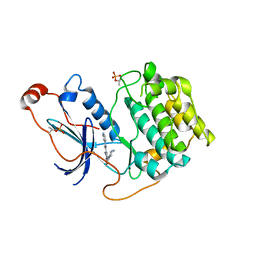 | | Crystal structure of PKCiota in complex with inhibitor | | Descriptor: | 2-amino-5-[3-(piperazin-1-yl)phenyl]-N-(pyridin-4-yl)pyridine-3-carboxamide, Protein kinase C iota type | | Authors: | Baburajendran, N, Hill, J. | | Deposit date: | 2018-10-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.261 Å) | | Cite: | Fragment-based Discovery of a Small-Molecule Protein Kinase C-iota Inhibitor Binding Post-kinase Domain Residues.
Acs Med.Chem.Lett., 10, 2019
|
|
2B97
 
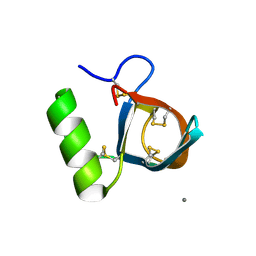 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3KSY
 
 | |
5CE7
 
 | | Structure of a non-canonical CID of Ctk3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CTD kinase subunit gamma | | Authors: | Muehlbacher, W, Mayer, A, Sun, M, Remmert, M, Cheung, A.C, Niesser, J, Soeding, J, Cramer, P. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ctk3, a subunit of the RNA polymerase II CTD kinase complex, reveals a noncanonical CTD-interacting domain fold.
Proteins, 83, 2015
|
|
5CEL
 
 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
4JCM
 
 | | Crystal structure of Gamma-CGTASE from Alkalophilic bacillus clarkii at 1.65 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, D, Zhou, J, Wu, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Gamma-Cgtase from Alkalophilic Bacillus Clarkii at 1.65 Angstrom Resolution.
To be Published
|
|
4ONQ
 
 | |
5CAJ
 
 | | Crystal structure of E. coli YaaA, a member of the DUF328/UPF0246 family | | Descriptor: | BENZAMIDINE, CHLORIDE ION, UPF0246 protein YaaA | | Authors: | Prahlad, J, Lin, J, Wilson, M.A. | | Deposit date: | 2015-06-29 | | Release date: | 2016-02-24 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The DUF328 family member YaaA is a DNA-binding protein with a novel fold.
J.Biol.Chem., 295, 2020
|
|
5CB2
 
 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
2X8E
 
 | | Discovery of a Novel Class of triazolones as Checkpoint Kinase Inhibitors - Hit to Lead Exploration | | Descriptor: | 5-METHYL-8-PYRIDIN-4-YL[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Novel Class of Triazolones as Checkpoint Kinase Inhibitors-Hit to Lead Exploration.
Bioorg.Med.Chem., 20, 2010
|
|
1ZZU
 
 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-2,4-L-Diaminobutyric Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
2X8I
 
 | | Discovery of a Novel Class of triazolones as Checkpoint Kinase Inhibitors - Hit to Lead Exploration | | Descriptor: | 7-(3-hydroxyphenyl)-5-methyl[1,2,4]triazolo[4,3-a]quinolin-1(2H)-one, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Otterbein, L, Vallentine, A, White, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of a Novel Class of Triazolones as Checkpoint Kinase Inhibitors-Hit to Lead Exploration.
Bioorg.Med.Chem., 20, 2010
|
|
3KWW
 
 | | Crystal structure of the 'restriction triad' mutant of HLA B*3508, beta-2-microglobulin and EBV peptide | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-01 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 2010
|
|
2Z3A
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
2Z8N
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, SULFATE ION | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5BYD
 
 | | Crystal structure of human ribokinase in P21 spacegroup | | Descriptor: | POTASSIUM ION, Ribokinase, SODIUM ION | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ribokinase in P21 spacegroup
To Be Published
|
|
5C0U
 
 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
6K0B
 
 | | cryo-EM structure of archaeal Ribonuclease P with mature tRNA | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
6K9C
 
 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
6KE1
 
 | | Crystal structure of TtCas1 | | Descriptor: | CRISPR-associated endonuclease Cas1 2 | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
3LJR
 
 | | GLUTATHIONE TRANSFERASE (THETA CLASS) FROM HUMAN IN COMPLEX WITH THE GLUTATHIONE CONJUGATE OF 1-MENAPHTHYL SULFATE | | Descriptor: | 1-MENAPHTHYL GLUTATHIONE CONJUGATE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
6GL2
 
 | | Structure of ZgEngAGH5_4 wild type at 1.2 Angstrom resolution | | Descriptor: | Endoglucanase, family GH5, IMIDAZOLE | | Authors: | Dorival, J, Ruppert, S, Gunnoo, M, Orlowski, A, Chapelais, M, Dabin, J, Labourel, A, Thompson, D, Michel, G, Czjzek, M, Genicot, S. | | Deposit date: | 2018-05-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Biochem. J., 475, 2018
|
|
