2BO4
 
 | | Dissection of mannosylglycerate synthase: an archetypal mannosyltransferase | | Descriptor: | CITRATE ANION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2YNV
 
 | | Cys221 oxidized, Mono zinc GIM-1 - GIM-1-Ox. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta- lactamases | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
2Y8R
 
 | | Crystal structure of apo AMA1 mutant (Tyr230Ala) from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
2YOP
 
 | | Long wavelength S-SAD structure of FAM3B PANDER | | Descriptor: | GLYCEROL, PROTEIN FAM3B | | Authors: | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
2YP3
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UCU
 
 | | STRUCTURAL AND MECHANISTIC INSIGHTS INTO HEMOGLOBIN-CATALYZED HYDROGEN SULFIDE OXIDATION AND THE FATE OF POLYSULFIDE PRODUCTS | | Descriptor: | HYDROSULFURIC ACID, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Vitvitsky, V, Yadav, P.K, An, S, Seravalli, J, Cho, U.-S, Banerjee, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural and Mechanistic Insights into Hemoglobin-catalyzed Hydrogen Sulfide Oxidation and the Fate of Polysulfide Products.
J. Biol. Chem., 292, 2017
|
|
2YI1
 
 | | Crystal structure of N-Acetylmannosamine kinase in complex with N- acetyl mannosamine 6-phosphate and ADP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
5UCP
 
 | | Class II fructose-1,6-bisphosphate aldolase E142A variant of Helicobacter pylori with FBP and cleavage products | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
5UD3
 
 | | Class II fructose-1,6-bisphosphate aldolase H180Q variant of Helicobacter pylori with FBP | | Descriptor: | 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
5UD0
 
 | | Class II fructose-1,6-bisphosphate aldolase E149A variant of Helicobacter pylori with cleavage products | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, CALCIUM ION, Fructose-bisphosphate aldolase, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
2Y64
 
 | | Xylopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y7C
 
 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
2YBD
 
 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
6MX9
 
 | | Lysozyme bound to 3-Aminophenol | | Descriptor: | 1,2-ETHANEDIOL, 3-aminophenol, BENZAMIDINE, ... | | Authors: | Blackburn, A, Partowmah, S.H, Brennan, H.M, Mestizo, K.E, Stivala, C.D, Petreczky, J, Perez, A, Horn, A, McSweeney, S. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A simple technique to improve microcrystals using gel exclusion of nucleation inducing elements
To Be Published
|
|
1PXV
 
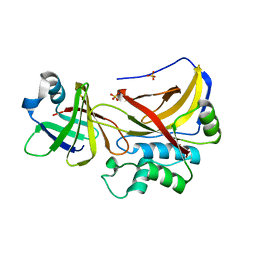 | | The staphostatin-staphopain complex: a forward binding inhibitor in complex with its target cysteine protease | | Descriptor: | GUANIDINE, SULFATE ION, cysteine protease, ... | | Authors: | Filipek, R, Rzychon, M, Oleksy, A, Gruca, M, Dubin, A, Potempa, J, Bochtler, M. | | Deposit date: | 2003-07-07 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Staphostatin-Staphopain Complex: A FORWARD BINDING INHIBITOR IN COMPLEX WITH ITS TARGET CYSTEINE PROTEASE.
J.Biol.Chem., 278, 2003
|
|
2Z5Y
 
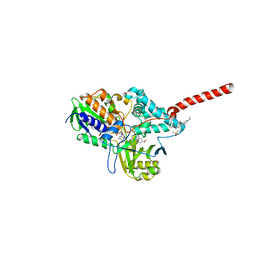 | | Crystal Structure of Human Monoamine Oxidase A (G110A) with Harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Amine oxidase [flavin-containing] A, DECYL(DIMETHYL)PHOSPHINE OXIDE, ... | | Authors: | Son, S.Y, Ma, J, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2007-07-20 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6MTE
 
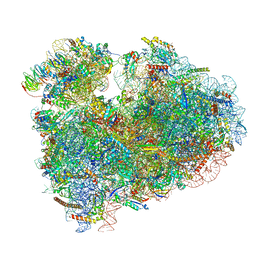 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
2Z6J
 
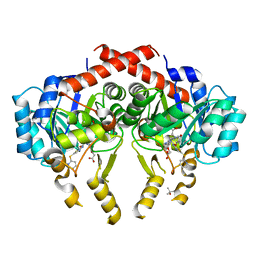 | | Crystal Structure of S. pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabK) in Complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-(2-((3-(5-(PYRIDIN-2-YLTHIO)THIAZOL-2-YL)UREIDO)METHYL)-1H-IMIDAZOL-4-YL)PHENOXY)ACETIC ACID, CALCIUM ION, ... | | Authors: | Saito, J, Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-08-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor.
Protein Sci., 17, 2008
|
|
6N3N
 
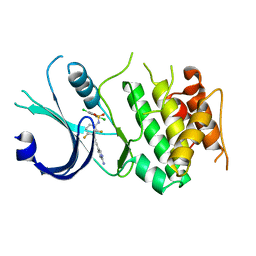 | | Identification of novel, potent and selective GCN2 inhibitors as first-in-class anti-tumor agents | | Descriptor: | N-{3-[(2-aminopyrimidin-5-yl)ethynyl]-2,4-difluorophenyl}-2,5-dichloro-3-(hydroxymethyl)benzene-1-sulfonamide, eIF-2-alpha kinase GCN2,eIF-2-alpha kinase GCN2 | | Authors: | Hoffman, I.D, Fujimoto, J, Kurasawa, O, Takagi, T, Klein, M.G, Kefala, G, Ding, S.C, Cary, D.R, Mizojiri, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Identification of Novel, Potent, and Orally Available GCN2 Inhibitors with Type I Half Binding Mode.
Acs Med.Chem.Lett., 10, 2019
|
|
1PYY
 
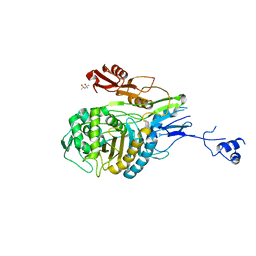 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
1K9S
 
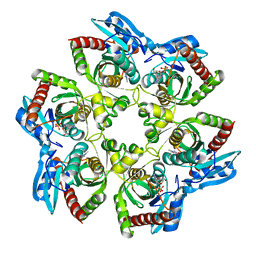 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | Descriptor: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | Authors: | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | Deposit date: | 2001-10-30 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
1PZZ
 
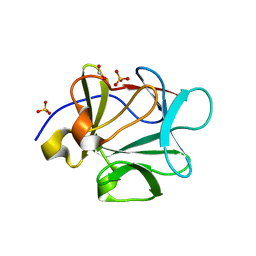 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
5UCN
 
 | | Class II fructose-1,6-bisphosphate aldolase E142A variant of Helicobacter pylori with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
6N9K
 
 | | Beta-lactamase from Escherichia coli str. Sakai | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Beta-lactamase from Escherichia coli str. Sakai
to be published
|
|
