4ZPY
 
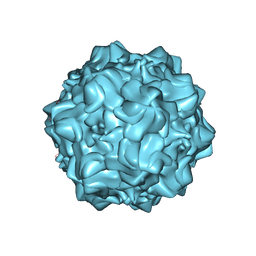 | | Structure of N170A MVM mutant empty capsid | | Descriptor: | VP1 protein | | Authors: | Guerra, P, Querol-Audi, J, Silva, C, Mateu, M.G, Verdaguer, N. | | Deposit date: | 2015-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for biologically relevant mechanical stiffening of a virus capsid by cavity-creating or spacefilling mutations.
Sci Rep, 7, 2017
|
|
3TAI
 
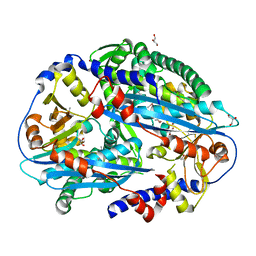 | | Crystal structure of NurA | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3GA2
 
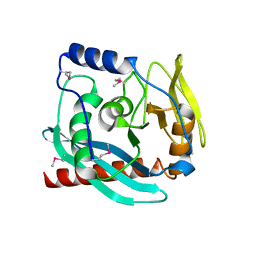 | | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624 | | Descriptor: | Endonuclease V | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owens, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624
To be Published
|
|
3TCA
 
 | |
5H7O
 
 | | Crystal structure of DJ-101 in complex with tubulin protein | | Descriptor: | 2-(1H-indol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Arnst, K, Wang, Y, Hwang, D.-J, Xue, Y, Costello, T, Hamilton, D, Chen, Q, Yang, J, Park, F, Dalton, J.T, Miller, D.D, Li, W. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance.
Cancer Res., 78, 2018
|
|
2PZY
 
 | | Structure of MK2 Complexed with Compound 76 | | Descriptor: | (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE, MAP kinase-activated protein kinase 2, STAUROSPORINE | | Authors: | White, A, Wu, J.P, Wang, J, Abeywardane, A, Andersen, D, Emmanuel, M, Gautschi, E, Goldberg, D.R, Kashem, M.A, Lukas, S, Mao, W, Martin, L, Morwick, T, Moss, N, Pargellis, C, Patel, U.R, Patnaude, L, Peet, G.W, Skow, D, Snow, R.J, Ward, Y, Werneburg, B. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The discovery of carboline analogs as potent MAPKAP-K2 inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4ZEX
 
 | | Crystal structure of PfHAD1 in complex with glyceraldehyde-3-phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
2Q00
 
 | | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus. NESG target SsR10. | | Descriptor: | Orf c02003 protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Owens, L, Ma, L.-C, Cunningham, K, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus.
To be Published
|
|
5I1E
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV1-39 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
3TES
 
 | | Crystal Structure of Tencon | | Descriptor: | Tencon | | Authors: | Luo, J, Jacobs, S, Teplyakov, A, Obmolova, G, O'Neil, K, Gilliland, G. | | Deposit date: | 2011-08-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of novel FN3 domains with high stability by a consensus sequence approach.
Protein Eng.Des.Sel., 25, 2012
|
|
5I2H
 
 | | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, FORMIC ACID, ... | | Authors: | Chang, C, Duke, N, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-08 | | Release date: | 2016-03-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin.
To Be Published
|
|
3TFA
 
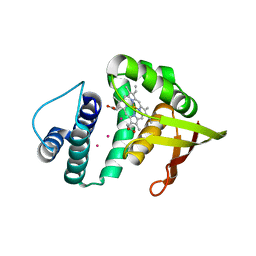 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 under 6 atm of xenon | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, XENON | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2711 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4ZJH
 
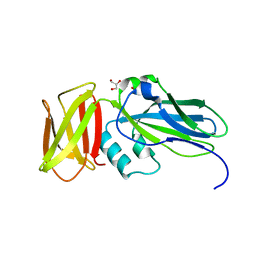 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains NIE-MG1. | | Descriptor: | ACETATE ION, GLYCEROL, alpha-2-Macroglobulin | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3GDS
 
 | |
5I08
 
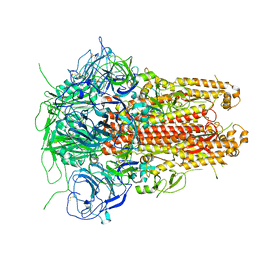 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
5I4C
 
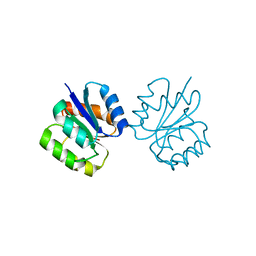 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | Descriptor: | Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
5I1H
 
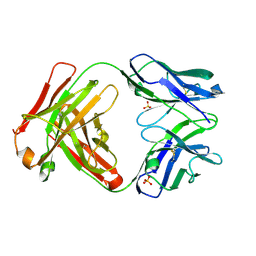 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
3GEP
 
 | |
6T2G
 
 | |
6TEY
 
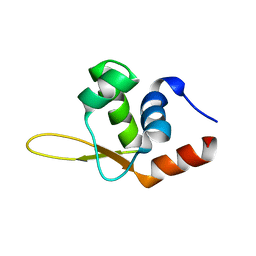 | |
5I18
 
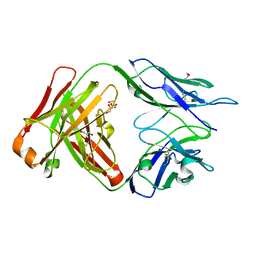 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
6TRM
 
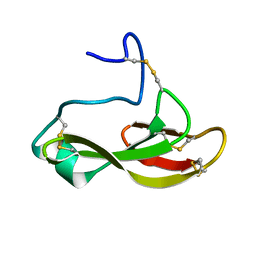 | | Solution structure of the antifungal protein PAFC | | Descriptor: | Pc21g12970 protein | | Authors: | Czajlik, A, Holzknecht, J, Marx, F, Batta, G. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Int J Mol Sci, 22, 2021
|
|
3GFU
 
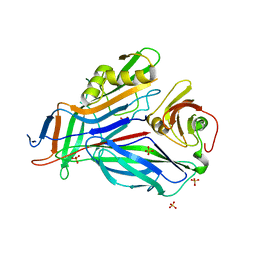 | | FaeE-FaeG chaperone-major pilin complex of F4 ac 5/95 fimbriae | | Descriptor: | Chaperone protein faeE, FaeG, SULFATE ION | | Authors: | Van Molle, I, Moonens, K, Garcia-Pino, A, Buts, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2009-02-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
J.Mol.Biol., 394, 2009
|
|
5I1K
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV3-20 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
