7X62
 
 | | Crystal structure of AtHPPD-Y191051 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X6M
 
 | | Crystal structure of AtHPPD-Y191058 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-2-(trifluoromethyl)quinazolin-4-one, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X5U
 
 | | Crystal structure of AtHPPD-diketonitrile complex | | Descriptor: | (1R,2S,3R)-1-cyclopropyl-2-(iminomethyl)-3-[2-methylsulfonyl-4-(trifluoromethyl)phenyl]propane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X64
 
 | | Crystal structure of AtHPPD-Y18027 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X6N
 
 | | Crystal structure of AtHPPD-Y191710 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-4-oxidanylidene-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2-carboxylate | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X8I
 
 | | Crystal structure of AtHPPD-Shikonin complex | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
2NW8
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and tryptophan. Northeast Structural Genomics Target XcR13. | | Descriptor: | MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1YRB
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
8DK6
 
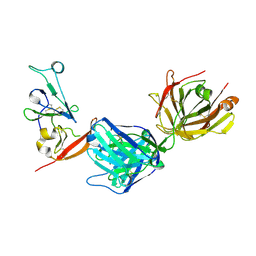 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
5YBV
 
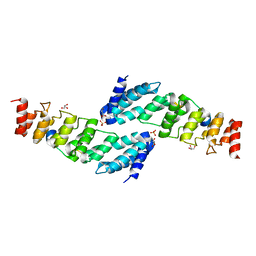 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
4FQP
 
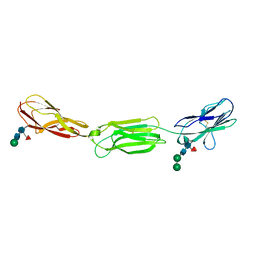 | | Crystal structure of human Nectin-like 5 full ectodomain (D1-D3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, ... | | Authors: | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6MOP
 
 | |
6MPK
 
 | |
3ZF3
 
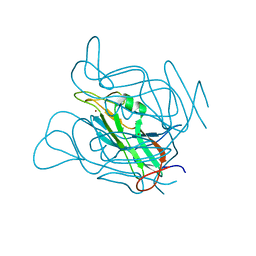 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84I mutant). | | Descriptor: | DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
1YR8
 
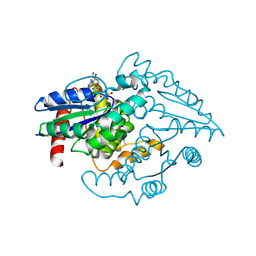 | | PAB0955 crystal structure : a GTPase in GTP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
6MQY
 
 | |
1YFV
 
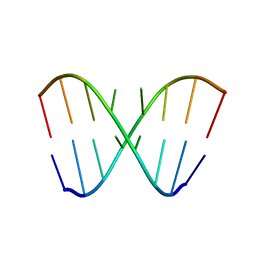 | |
4GS6
 
 | | Irreversible Inhibition of TAK1 Kinase by 5Z-7-Oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, 1,2-ETHANEDIOL, Tak1-Tab1 fusion protein | | Authors: | Larsen, N.A, Ferguson, A.D, Wu, J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-01-23 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism and In Vitro Pharmacology of TAK1 Inhibition by (5Z)-7-Oxozeaenol.
Acs Chem.Biol., 8, 2013
|
|
5YBU
 
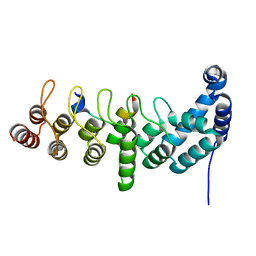 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YNX
 
 | | Structure of house dust mite allergen Der f 21 in PEG400 | | Descriptor: | Allergen Der f 21, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I, Beis, K, Say, Y.H. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
3ZEZ
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
8D37
 
 | |
1K6X
 
 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
8D33
 
 | |
8D3R
 
 | |
