8VBT
 
 | |
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
5JFL
 
 | |
2IBO
 
 | | X-ray Crystal Structure of Protein SP2199 from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR31 | | Descriptor: | Hypothetical protein SP2199 | | Authors: | Seetharaman, J, Abashidze, M, Forouhar, F, Shastry, R, Conover, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein SP2199 from Streptococcus pneumoniae, Northeast structural genomics target SpR31
To be Published
|
|
2OR4
 
 | | A high resolution crystal structure of human glutamate carboxypeptidase II in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Lubkowski, J. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insight into the pharmacophore pocket of human glutamate carboxypeptidase II.
J.Med.Chem., 50, 2007
|
|
8DLA
 
 | | ClpP2 from Chlamydia trachomatis bound by MAS1-12 | | Descriptor: | 1-{4-[(4-chlorophenyl)methyl]piperazin-1-yl}-2-methyl-2-[5-(trifluoromethyl)pyridine-2-sulfonyl]propan-1-one, ATP-dependent Clp protease proteolytic subunit 2, GLYCEROL, ... | | Authors: | Azadmanesh, J, Struble, L.R, Seleem, M.A, Ouellette, S, Conda-Sheridan, M, Borgstahl, G.E.O. | | Deposit date: | 2022-07-07 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The structure of caseinolytic protease subunit ClpP2 reveals a functional model of the caseinolytic protease system from Chlamydia trachomatis.
J.Biol.Chem., 299, 2023
|
|
2I81
 
 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
2PML
 
 | |
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
5WY5
 
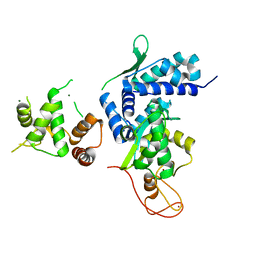 | | Crystal structure of MAGEG1 and NSE1 complex | | Descriptor: | MAGNESIUM ION, Melanoma-associated antigen G1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Yang, M, Gao, J. | | Deposit date: | 2017-01-11 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Mage-Ring Protein Complexes Comprise A Family Of E3 Ubiquitin Ligases.
Mol.Cell, 39, 2010
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
5WZG
 
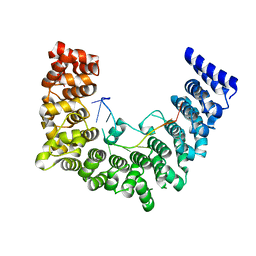 | | Structure of APUM23-GAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
6MQJ
 
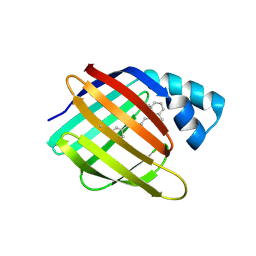 | |
7QIR
 
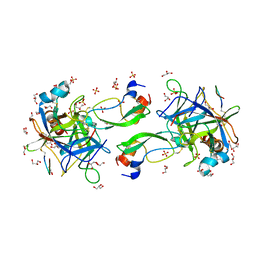 | | CRYSTAL STRUCTURE OF THE P1 monofluorethylglycine(MfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
5JNP
 
 | | Crystal structure of a rice (Oryza Sativa) cellulose synthase plant conserved region (P-CR) | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Rushton, P.S, Olek, A.T, Makowski, L, Badger, J, Steussy, C.N, Carpita, N.C, Stauffacher, C.V. | | Deposit date: | 2016-04-30 | | Release date: | 2016-12-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Rice Cellulose SynthaseA8 Plant-Conserved Region Is a Coiled-Coil at the Catalytic Core Entrance.
Plant Physiol., 173, 2017
|
|
3TFM
 
 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
2PL9
 
 | |
7QIS
 
 | | CRYSTAL STRUCTURE OF THE P1 difluoroethylglycine (DfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
5WBM
 
 | |
2HOH
 
 | | RIBONUCLEASE T1 (N9A MUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
5IG5
 
 | |
5W6A
 
 | | HLA-C*06:02 presenting ARTELYRSL | | Descriptor: | ALA-ARG-THR-GLU-LEU-TYR-ARG-SER-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
8EAP
 
 | |
5IHW
 
 | | The crystal structure of SdrE from staphylococcus aureus | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
