5IZA
 
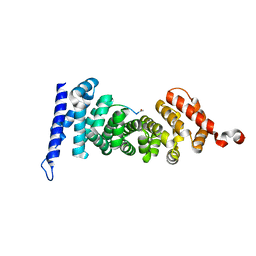 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4RK1
 
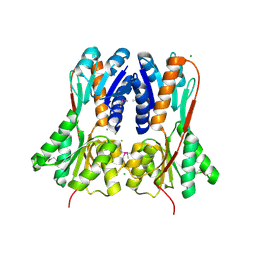 | | Crystal structure of LacI family transcriptional regulator from Enterococcus faecium, Target EFI-512930, with bound ribose | | Descriptor: | CHLORIDE ION, Ribose transcriptional regulator, alpha-D-ribofuranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator Rbsr from Enterococcus faecium, Target EFI-512930
To be Published
|
|
6Q2O
 
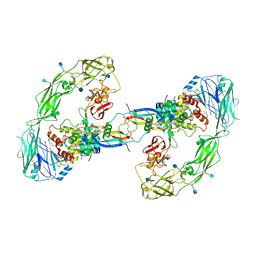 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
5IW4
 
 | |
4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
4RXL
 
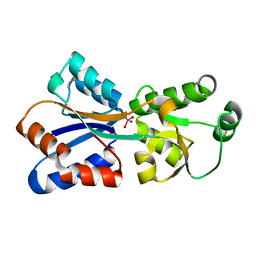 | | Crystal structure of Molybdenum ABC transporter solute binding protein Vc_A0726 from Vibrio Cholerae, Target EFI-510913, in complex with tungstate | | Descriptor: | Molybdenum ABC transporter, periplasmic molybdenum-binding protein, TUNGSTATE(VI)ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Molybdate Transporter Vc_A0726 from Vibrio Cholerae, Target EFI-510913
To be Published
|
|
4RY9
 
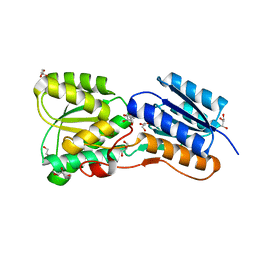 | | Crystal structure of carbohydrate transporter solute binding protein VEIS_2079 from Verminephrobacter eiseniae EF01-2, TARGET EFI-511009, a complex with D-TALITOL | | Descriptor: | BICARBONATE ION, D-altritol, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of carbohydrate transporter solute binding protein VEIS_2079 from Verminephrobacter eiseniae
To be Published
|
|
6MRK
 
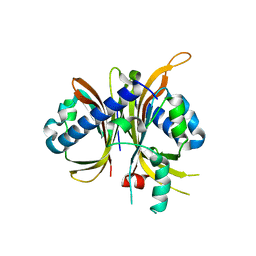 | |
7SZE
 
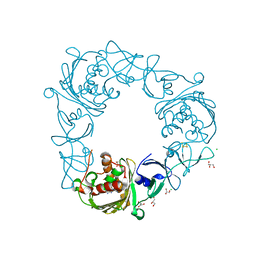 | |
4RYA
 
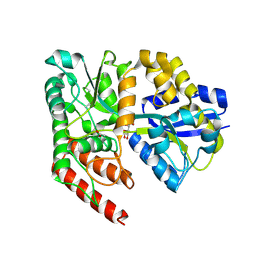 | | Crystal structure of abc transporter solute binding protein AVI_3567 from AGROBACTERIUM VITIS S4, TARGET EFI-510645, with bound D-mannitol | | Descriptor: | ABC transporter substrate binding protein (Sorbitol), ACETATE ION, D-MANNITOL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Periplasmic Solute-Binding Protein Avi_3567 from Agrobacterium Vitis, Target Efi-510645
To be Published
|
|
7SZH
 
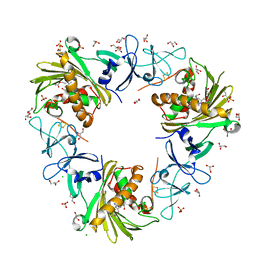 | |
7SZG
 
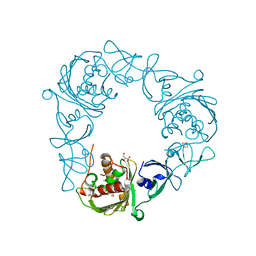 | | Structure of the Rieske Non-heme Iron Oxygenase GxtA Pressurized with Xenon | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Bridwell-Rabb, J, Liu, J. | | Deposit date: | 2021-11-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design principles for site-selective hydroxylation by a Rieske oxygenase.
Nat Commun, 13, 2022
|
|
7SZF
 
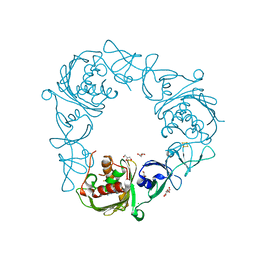 | |
4S3C
 
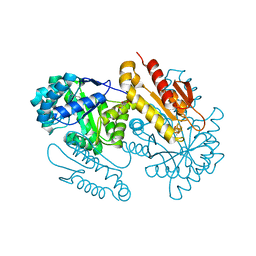 | | IspG in complex with Epoxide Intermediate | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6Q9H
 
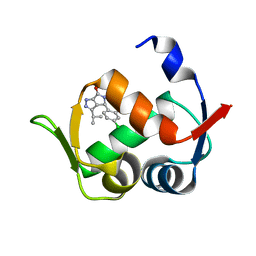 | |
5JA5
 
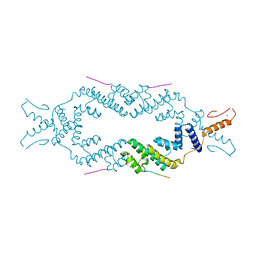 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
6Q9Y
 
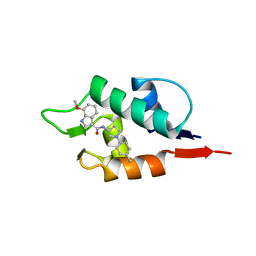 | |
4TRG
 
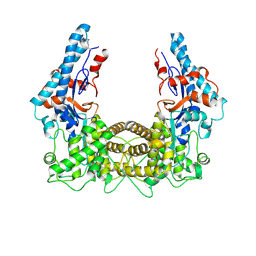 | | the SNL domain of SidC | | Descriptor: | MERCURY (II) ION, SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
4RTT
 
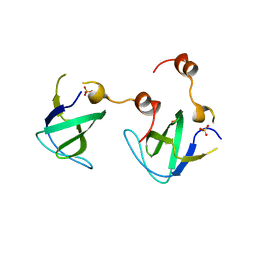 | |
7SPA
 
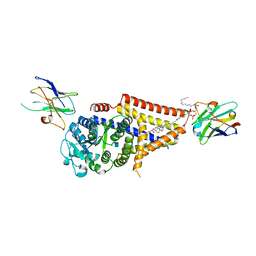 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
5K8Y
 
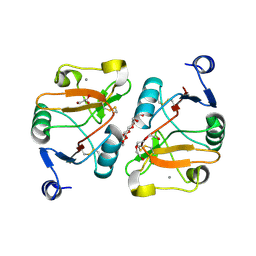 | | Structure of the Mus musclus Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
8CJB
 
 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7SP9
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP6
 
 | | Chlorella virus hyaluronan synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
