3DMD
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
4AM0
 
 | | Structure of Dengue virus strain 4 DIII in complex with Fab 2H12 | | Descriptor: | ENVELOPE PROTEIN,, FAB 2H12, HEAVY CHAIN, ... | | Authors: | Midgley, C.M, Flanagan, A, Mongkolsapaya, J, Grimes, J.M, Screaton, G.R. | | Deposit date: | 2012-03-06 | | Release date: | 2012-06-06 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Analysis of a Dengue Cross-Reactive Antibody Complexed with Envelope Domain III Reveals the Molecular Basis of Cross-Reactivity.
J.Immunol., 188, 2012
|
|
6KJW
 
 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
1JSL
 
 | | Crystal structure of Erwinia chrysanthemi L-asparaginase complexed with 6-HYDROXY-D-NORLEUCINE | | Descriptor: | 6-HYDROXY-D-NORLEUCINE, GLYCEROL, L-asparaginase, ... | | Authors: | Aghaiypour, K, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2001-08-17 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Do bacterial L-asparaginases utilize a catalytic triad Thr-Tyr-Glu?
Biochim.Biophys.Acta, 1550, 2001
|
|
4A5V
 
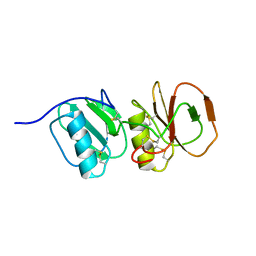 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
4AR2
 
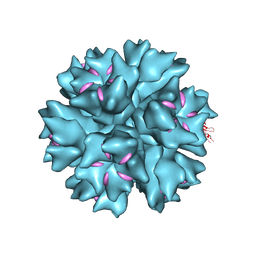 | | Dodecahedron formed of penton base protein from adenovirus Ad3 | | Descriptor: | CALCIUM ION, FIBER PROTEIN, L2 PROTEIN III (PENTON BASE) | | Authors: | Burmeister, W.P, Szolajska, E, Zochowska, M, Nerlo, B, Andreev, I, Schoehn, G, Andrieu, J.-P, Fender, P, Naskalska, A, Zubieta, C, Cusack, S, Chroboczek, J. | | Deposit date: | 2012-04-20 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis for the Integrity of Adenovirus Ad3 Dodecahedron.
Plos One, 7, 2012
|
|
6C0V
 
 | |
4AFK
 
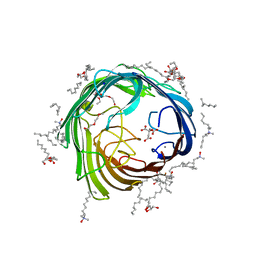 | | In meso structure of alginate transporter, AlgE, from Pseudomonas aeruginosa, PAO1 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Tan, J, Pye, V.E, Aragao, D, Caffrey, M. | | Deposit date: | 2012-01-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Conformational Landscape for Alginate Secretion Across the Outer Membrane of Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3DOL
 
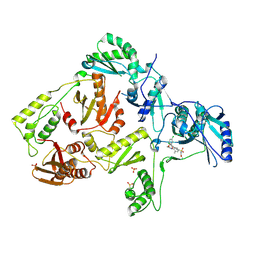 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
4AKA
 
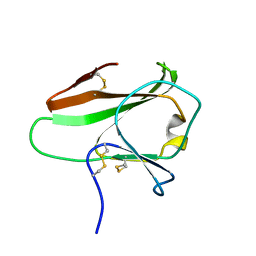 | | IPSE alpha-1, an IgE-binding crystallin | | Descriptor: | IL-4-INDUCING PROTEIN | | Authors: | Meyer, N.H, Mayerhofer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Mueller-Dieckmann, J, Sattler, M, Scharmm, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Crystallin Fold in the Interleukin-4-Inducing Principle of Schistosoma Mansoni Eggs (Ipse/Alpha-1) Mediates Ige Binding for Antigen-Independent Basophil Activation
J.Biol.Chem., 290, 2015
|
|
6BW6
 
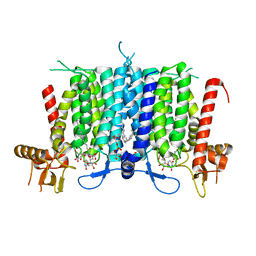 | | Human GPT (DPAGT1) H129 variant in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3DPN
 
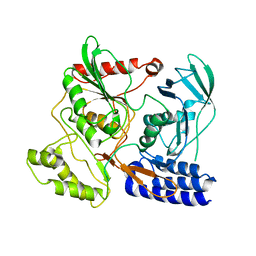 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3IJT
 
 | | Structural Characterization of SMU.440, a Hypothetical Protein from Streptococcus mutans | | Descriptor: | Putative uncharacterized protein | | Authors: | Nan, J, Brostromer, E, Kristensen, O, Su, X.-D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance
Plos One, 4, 2009
|
|
6BXB
 
 | | Crystal structure of an extended b3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4A06
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Activator PS114 Bound to the PIF-Pocket | | Descriptor: | (3S)-4-(5-chloro-1H-benzimidazol-2-yl)-3-(4-chlorophenyl)butanoic acid, 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schulze, J.O, Lopez-Garcia, L.A, Froehner, W, Zhang, H, Navratil, J, Hindie, V, Zeuzem, S, Alzari, P.M, Neimanis, S, Engel, M, Biondi, R.M. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Protein Kinase Pkczeta by the N-Terminal C1 Domain and Small Compounds to the Pif-Pocket.
Chem.Biol., 18, 2011
|
|
6K1B
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | Descriptor: | Gamma-aminobutyraldehyde dehydrogenase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-11 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
2NYR
 
 | | Crystal Structure of Human Sirtuin Homolog 5 in Complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, NAD-dependent deacetylase sirtuin-5, ZINC ION | | Authors: | Min, J.R, Antoshenko, T, Allali-Hassani, A, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin.
Structure, 15, 2007
|
|
6K43
 
 | |
3ISV
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion
To be Published
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
3IU2
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096 | | Descriptor: | (2R)-2-{4-hydroxy-5-methoxy-2-[3-(4-methylpiperazin-1-yl)propyl]phenyl}-3-pyridin-3-yl-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096
To be Published
|
|
6DU8
 
 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
1GBY
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
