5VNH
 
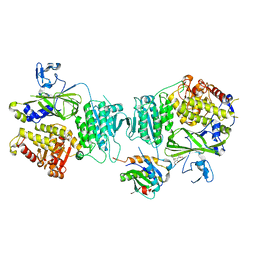 | |
3CGY
 
 | | Crystal Structure of Salmonella Sensor Kinase PhoQ catalytic domain in complex with radicicol | | Descriptor: | RADICICOL, Virulence sensor histidine kinase phoQ | | Authors: | Guarnieri, M.T, Zhang, L, Shen, J, Zhao, R. | | Deposit date: | 2008-03-06 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Hsp90 inhibitor radicicol interacts with the ATP-binding pocket of bacterial sensor kinase PhoQ.
J.Mol.Biol., 379, 2008
|
|
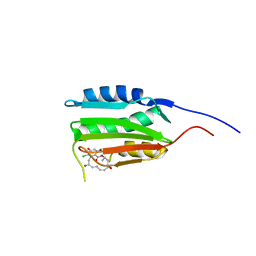
3D7S
 
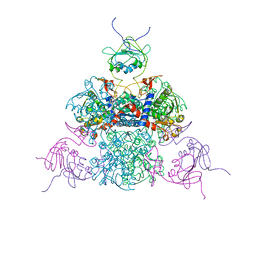 | |
5VXQ
 
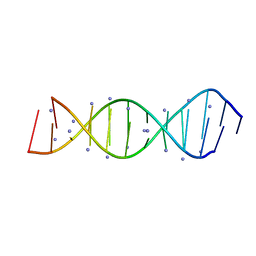 | | X-Ray crystallography structure of the parallel stranded duplex formed by 5-rA5-dA-rA5 | | Descriptor: | AMMONIUM ION, DNA/RNA (5'-R(*AP*AP*AP*AP*A)-D(P*A)-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Xie, J, Chen, Y, Wei, X, Kozlov, G, Gehring, K. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Influence of nucleotide modifications at the C2' position on the Hoogsteen base-paired parallel-stranded duplex of poly(A) RNA.
Nucleic Acids Res., 45, 2017
|
|
6ISC
 
 | | complex structure of mCD226-ecto and hCD155-D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
6BW5
 
 | | Human GPT (DPAGT1) in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3F2C
 
 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP and Mn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4C4Z
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Y
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A4 | | Descriptor: | 1-(3-chlorophenyl)-3-(2-methoxyethyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4X
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3IP8
 
 | | Crystal structure of arylmalonate decarboxylase (AMDase) from Bordatella bronchiseptic in complex with benzylphosphonate | | Descriptor: | Arylmalonate decarboxylase, benzylphosphonic acid | | Authors: | Okrasa, K, Levy, C, Leys, D, Micklefield, J. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure-Guided Directed Evolution of Alkenyl and Arylmalonate Decarboxylases.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5W0M
 
 | | Structure of human TUT7 catalytic module (CM) in complex with U5 RNA | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3F0X
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-27 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
3IU1
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA
To be Published
|
|
1K03
 
 | | Crystal Structure of Old Yellow Enzyme Mutant Gln114Asn Complexed with Para-hydroxy Benzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH DEHYDROGENASE 1, ... | | Authors: | Brown, B.J, Hyun, J, Duvvuri, S.D, Karplus, P.A, Massey, V. | | Deposit date: | 2001-09-18 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of glutamine 114 in old yellow enzyme
J.Biol.Chem., 277, 2002
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
3IQF
 
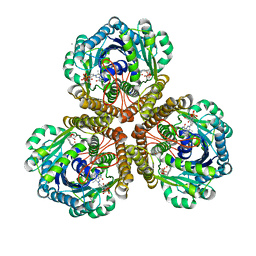 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methenyl-tetrahydromethanopterin | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, CALCIUM ION, F420-dependent methylenetetrahydromethanopterin dehydrogenase, ... | | Authors: | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2009-08-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
3EY0
 
 | | A new form of DNA-drug interaction in the minor groove of a coiled coil | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, 5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DT)-3', MAGNESIUM ION | | Authors: | Pous, J, Moreno, T, Subirana, J.A, Campos, J.L. | | Deposit date: | 2008-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Coiled-coil conformation of a pentamidine-DNA complex
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3IS5
 
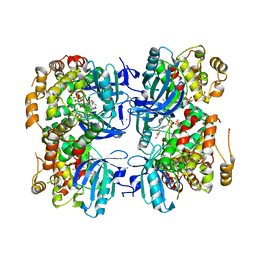 | | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Senisterra, G, MacKenzie, F, Hutchinson, A, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720
To be Published
|
|
1CTI
 
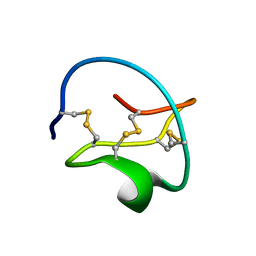 | |
3F1R
 
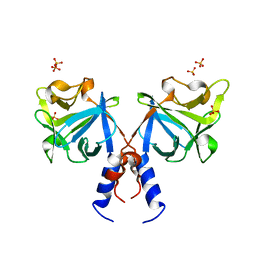 | | Crystal structure of FGF20 dimer | | Descriptor: | Fibroblast growth factor 20, SULFATE ION | | Authors: | Kalinina, J, Mohammadi, M. | | Deposit date: | 2008-10-28 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homodimerization controls the fibroblast growth factor 9 subfamily's receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix
Mol.Cell.Biol., 29, 2009
|
|
1JQG
 
 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
3EY5
 
 | | Putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron. | | Descriptor: | Acetyltransferase-like, GNAT family, SULFATE ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron.
To be Published
|
|
6HBK
 
 | | Echovirus 18 Open particle without one pentamer | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
