1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
6I6T
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-oxidanyl-naphthalene-2-carboxylic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I7X
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT53 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with RT53
To Be Published
|
|
3CP7
 
 | |
3IDC
 
 | | Crystal structure of (102-265)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
6I81
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT56
To Be Published
|
|
5T6W
 
 | | HLA-B*57:01 presenting SSTRGISQLW | | Descriptor: | Beta-2-microglobulin, Decapeptide: SER-SER-THR-ARG-GLY-ILE-SER-GLN-LEU-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T72
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
6BSS
 
 | | JAK2 JH2 in complex with NU6102 | | Descriptor: | GLYCEROL, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE, Tyrosine-protein kinase JAK2 | | Authors: | Puleo, D.E, Schlessinger, J. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | JAK2 JH2 Binders
To Be Published
|
|
3CQ5
 
 | | Histidinol-phosphate aminotransferase from Corynebacterium glutamicum in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Histidinol-phosphate aminotransferase, SULFATE ION, ... | | Authors: | Sandalova, T, Marienhagen, J, Schneider, G. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the structural basis of substrate recognition by histidinol-phosphate aminotransferase from Corynebacterium glutamicum
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4AEL
 
 | | PDE10A in complex with the inhibitor AZ5 | | Descriptor: | 2-(2'-ETHOXYBIPHENYL-4-YL)-4-HYDROXY-1,6-NAPHTHYRIDINE-3-CARBONITRILE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Bauer, U, Giordanetto, F, Bauer, M, OMahony, G, Johansson, K.E, Knecht, W, Hartleib-Geschwindner, J, Toppner Carlsson, E, Enroth, C, Sjogren, T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 4-Hydroxy-1,6-Naphthyridine-3-Carbonitrile Derivatives as Novel Pde10A Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6IAX
 
 | | MloK1 model from single particle analysis of 2D crystals, class 1 (extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6BWT
 
 | | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, SULFATE ION, Thioredoxin reductase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis.
To Be Published
|
|
4AFR
 
 | | THE STRUCTURE OF METACASPASE 2 (C213A MUTANT) FROM T. BRUCEI | | Descriptor: | METACASPASE | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3CQ4
 
 | |
1JCD
 
 | | Crystal Structure of a Novel Alanine-Zipper Trimer at 1.3 A Resolution, I6A,L9A,V13A,L16A,V20A,L23A,V27A,M30A,V34A,L48A,M51A mutations | | Descriptor: | MAJOR OUTER MEMBRANE LIPOPROTEIN | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An Alanine-Zipper Structure
Determined by Long Range
Intermolecular Interactions
J.Biol.Chem., 277, 2002
|
|
6IC6
 
 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
3IEP
 
 | | Firefly luciferase apo structure (P41 form) | | Descriptor: | Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
6BXF
 
 | | Crystal structure of an extended b3 integrin L33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
3IEZ
 
 | | Crystal structure of the RasGAP C-terminal (RGC) domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-23 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the RasGAP C-terminal (RGC) domain
of IQGAP2
To be Published
|
|
3CWQ
 
 | | Crystal structure of chromosome partitioning protein (ParA) in complex with ADP from Synechocystis sp. Northeast Structural Genomics Consortium Target SgR89 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ParA family chromosome partitioning protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Xiao, R, Janjua, H, Maglaqui, M, Foote, E.L, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of chromosome partitioning protein (ParA) in complex with ADP from Synechocystis sp.
To be Published
|
|
6IDL
 
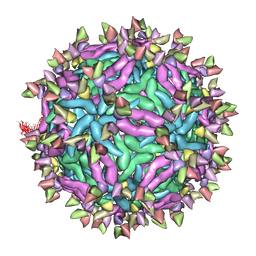 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class II particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
4AQQ
 
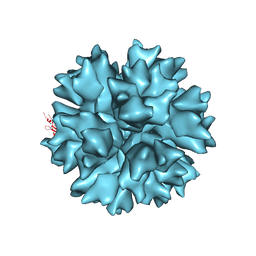 | | Dodecahedron formed of penton base protein from adenovirus Ad3 | | Descriptor: | CALCIUM ION, L2 PROTEIN III (PENTON BASE) | | Authors: | Burmeister, W.P, Szolajska, E, Zochowska, M, Nerlo, B, Andreev, I, Schoehn, G, Andrieu, J.-P, Fender, P, Naskalska, A, Zubieta, C, Cusack, S, Chroboczek, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | The Structural Basis for the Integrity of Adenovirus Ad3 Dodecahedron.
Plos One, 7, 2012
|
|
