4B2N
 
 | | Latex Oxygenase RoxA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 70 KDA PROTEIN, HEME C, ... | | Authors: | Seidel, J, Schmitt, G, Hoffmann, M, Jendrossek, D, Einsle, O. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the processive rubber oxygenase RoxA from Xanthomonas sp.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4B7R
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4B4R
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group B type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4B9Z
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with Acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ALPHA-GLUCOSIDASE, ... | | Authors: | Larsbrink, J, Izumi, A, Hemsworth, G.R, Davies, G.J, Brumer, H. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Enzymology of Cellvibrio Japonicus Agd31B Reveals Alpha-Transglucosylase Activity in Glycoside Hydrolase Family 31
J.Biol.Chem., 287, 2012
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4BDF
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 5-METHYL-3-PHENYL-1H-PYRAZOLE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4AV3
 
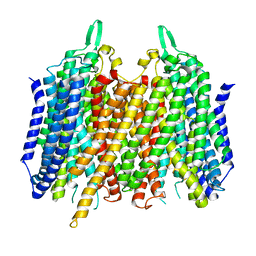 | | Crystal structure of Thermotoga Maritima sodium pumping membrane integral pyrophosphatase with metal ions in active site | | Descriptor: | CALCIUM ION, K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION | | Authors: | Kajander, T, Kogan, K, Kellosalo, J, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
4B2J
 
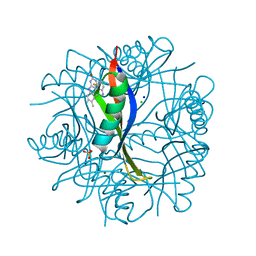 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Manipulation of a Flavoprotein Photocycle.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4ATY
 
 | |
1NB7
 
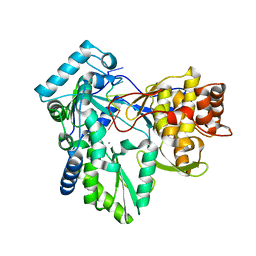 | | HC-J4 RNA polymerase complexed with short RNA template strand | | Descriptor: | 5'-R(*UP*UP*UP*U)-3', MANGANESE (II) ION, polyprotein | | Authors: | O'Farrell, D.J, Trowbridge, R, Rowlands, D.J, Jaeger, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation.
J.Mol.Biol., 326, 2003
|
|
1RYP
 
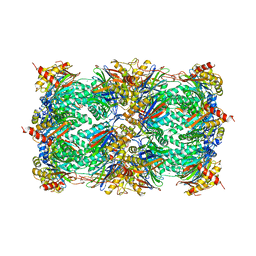 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
4B0Z
 
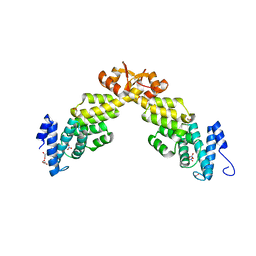 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
4N9O
 
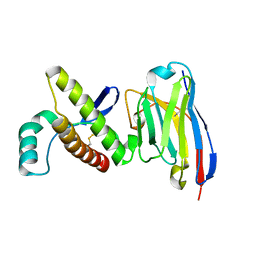 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4B5S
 
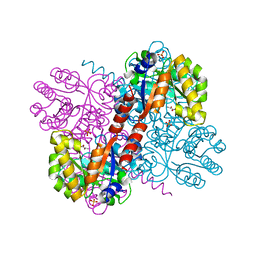 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, D-Glyceraldehyde, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4BA0
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with 5F-alpha-GlcF | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-beta-D-glucopyranose, ALPHA-GLUCOSIDASE, ... | | Authors: | Larsbrink, J, Izumi, A, Hemsworth, G.R, Davies, G.J, Brumer, H. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Enzymology of Cellvibrio Japonicus Agd31B Reveals Alpha-Transglucosylase Activity in Glycoside Hydrolase Family 31
J.Biol.Chem., 287, 2012
|
|
4NAZ
 
 | | Crystal Structure of FosB from Staphylococcus aureus with Zn and Sulfate at 1.15 Angstrom Resolution | | Descriptor: | GLYCEROL, Metallothiol transferase FosB, SULFATE ION, ... | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
1NDT
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (NITRITE REDUCTASE) | | Authors: | Dodd, F.E, Vanbeeumen, J, Eady, R.R, Hasnain, S.S. | | Deposit date: | 1998-10-28 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner.
J.Mol.Biol., 282, 1998
|
|
4ATN
 
 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, RIBOSOMAL RNA LARGE SUBUNIT METHYLTRANSFERASE M, ... | | Authors: | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
4B5Q
 
 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
4AWU
 
 | |
4B7J
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4AUD
 
 | |
1TCA
 
 | |
4AX1
 
 | | Q157N mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4B9Y
 
 | | Crystal Structure of Apo Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Izumi, A, Hemsworth, G.R, Davies, G.J, Brumer, H. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Enzymology of Cellvibrio Japonicus Agd31B Reveals Alpha-Transglucosylase Activity in Glycoside Hydrolase Family 31
J.Biol.Chem., 287, 2012
|
|
