2Q82
 
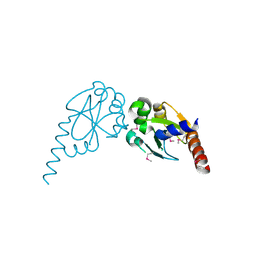 | | Crystal structure of core protein P7 from Pseudomonas phage phi12. Northeast Structural Genomics Target OC1 | | 分子名称: | Core protein P7 | | 著者 | Benach, J, Eryilmaz, E, Su, M, Seetharaman, J, Wei, H, Gottlieb, P, Hunt, J.F, Ghose, R, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-08 | | 公開日 | 2007-08-07 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structure and dynamics of the P7 protein from the bacteriophage phi 12.
J.Mol.Biol., 382, 2008
|
|
2Q5J
 
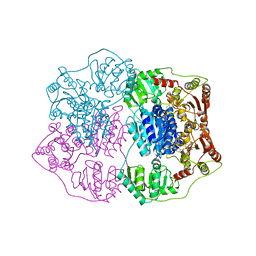 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Phenylpyruvate decarboxylase | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5O
 
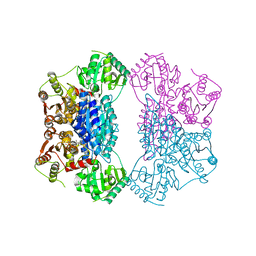 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP and phenylpyruvate | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, 3-PHENYLPYRUVIC ACID, GLYCEROL, ... | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2PLC
 
 | |
6PA5
 
 | |
7S8U
 
 | |
2F9C
 
 | | Crystal structure of YDCK from Salmonella cholerae. NESG target SCR6 | | 分子名称: | CESIUM ION, Hypothetical protein YDCK | | 著者 | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, H, Cooper, B, Acton, X.T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-12-05 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of YDCK from Salmonella cholerae NESG target SCR6.
To be Published
|
|
1SRA
 
 | | STRUCTURE OF A NOVEL EXTRACELLULAR CA2+-BINDING MODULE IN BM-40(SLASH)SPARC(SLASH)OSTEONECTIN | | 分子名称: | CALCIUM ION, SPARC | | 著者 | Hohenester, E, Maurer, P, Hohenadl, C, Timpl, R, Jansonius, J.N, Engel, J. | | 登録日 | 1995-08-21 | | 公開日 | 1996-03-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a novel extracellular Ca(2+)-binding module in BM-40.
Nat.Struct.Biol., 3, 1996
|
|
3T19
 
 | |
1SMK
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | 分子名称: | CITRIC ACID, Malate dehydrogenase, glyoxysomal | | 著者 | Cox, B, Chit, M.M, Weaver, T, Bailey, J, Gietl, C, Bell, E, Banaszak, L. | | 登録日 | 2004-03-09 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
Febs J., 272, 2005
|
|
6E6L
 
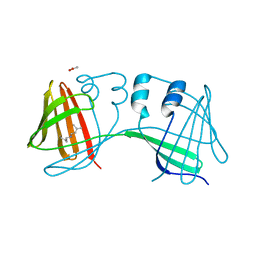 | |
6Y2M
 
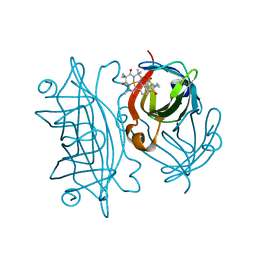 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | 分子名称: | Streptavidin, biotC4-1 cofactor | | 著者 | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | 登録日 | 2020-02-17 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
111D
 
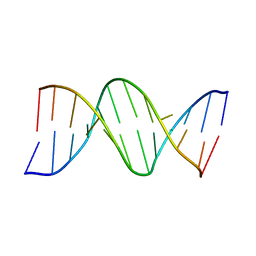 | |
4DZB
 
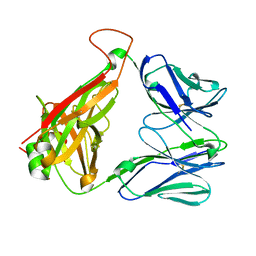 | |
6Y2T
 
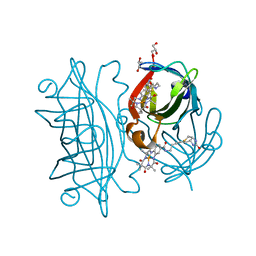 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | 分子名称: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | 著者 | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | 登録日 | 2020-02-17 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
3T3O
 
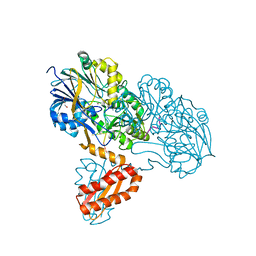 | | Molecular basis for the recognition and cleavage of RNA (CUGG) by the bifunctional 5'-3' exo/endoribonuclease RNase J | | 分子名称: | GLYCEROL, Metal dependent hydrolase, O2'methyl-RNA, ... | | 著者 | Dorleans, A, Li de la Sierra-Gallay, I, Piton, J, Zig, L, Gilet, L, Putzer, H, Condon, C. | | 登録日 | 2011-07-25 | | 公開日 | 2011-10-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular Basis for the Recognition and Cleavage of RNA by the Bifunctional 5'-3' Exo/Endoribonuclease RNase J.
Structure, 19, 2011
|
|
6Y3Q
 
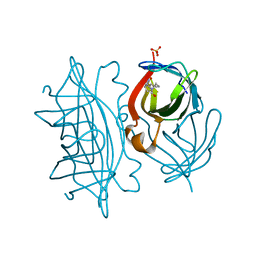 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | 分子名称: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | 著者 | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | 登録日 | 2020-02-18 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y9E
 
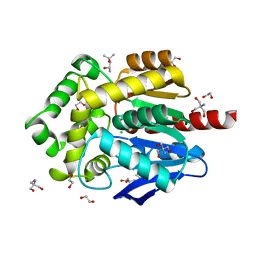 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chaloupkova, R, Damborsky, J, Marek, M. | | 登録日 | 2020-03-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
7SJ6
 
 | | T4 Lysozyme L99A/M102H with 1,2-Azaborine bound | | 分子名称: | 1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | 著者 | Yao, L, Wirth, J. | | 登録日 | 2021-10-16 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | T4 Lysozyme L99A/M102H with 1,2-Azaborine bound
to be published
|
|
3WL8
 
 | | Crystal Structure of pOPH S172A with octanoic acid | | 分子名称: | CITRIC ACID, OCTANOIC ACID (CAPRYLIC ACID), Oxidized polyvinyl alcohol hydrolase | | 著者 | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | 登録日 | 2013-11-08 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
7KIA
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | 分子名称: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | 著者 | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | 登録日 | 2020-10-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
1SHP
 
 | | THE NMR SOLUTION STRUCTURE OF A KUNITZ-TYPE PROTEINASE INHIBITOR FROM THE SEA ANEMONE STICHODACTYLA HELIANTHUS | | 分子名称: | TRYPSIN INHIBITOR | | 著者 | Antuch, W, Berndt, K, Chavez, M, Delfin, J, Wuthrich, K. | | 登録日 | 1992-11-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus.
Eur.J.Biochem., 212, 1993
|
|
7KIE
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 3 | | 分子名称: | CITRATE ANION, Fibroblast growth factor receptor 2, GLYCEROL, ... | | 著者 | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | 登録日 | 2020-10-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
4HK6
 
 | |
4E53
 
 | | Calmodulin and Nm peptide complex | | 分子名称: | Calmodulin, Linker, IQ motif of Neuromodulin | | 著者 | Kumar, V, Sivaraman, J. | | 登録日 | 2012-03-13 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
