8OZA
 
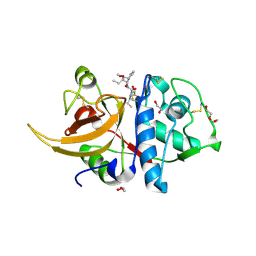 | | Human cathepsin L in complex with covalently bound CA-074 methyl ester | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | 著者 | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | 登録日 | 2023-05-08 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7SYW
 
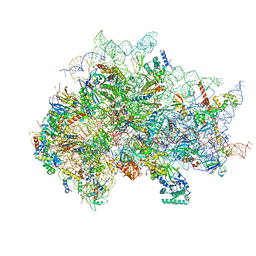 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6G4M
 
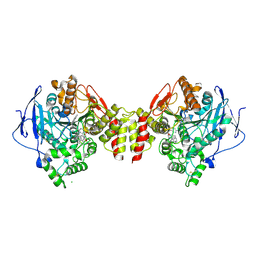 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 1 | | 分子名称: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | 登録日 | 2018-03-28 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYT
 
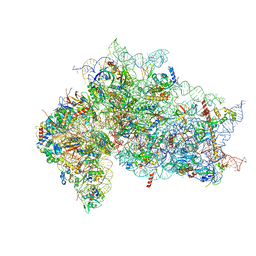 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6N3U
 
 | |
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-22 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TNW
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
3B3G
 
 | | The 2.4 A crystal structure of the apo catalytic domain of coactivator-associated arginine methyl transferase I(CARM1,140-480). | | 分子名称: | Histone-arginine methyltransferase CARM1 | | 著者 | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
1AM7
 
 | |
4PAD
 
 | |
2V90
 
 | | Crystal structure of the 3rd PDZ domain of intestine- and kidney- enriched PDZ domain IKEPP (PDZD3) | | 分子名称: | PDZ DOMAIN-CONTAINING PROTEIN 3, SULFATE ION | | 著者 | Uppenberg, J, Gileadi, C, Phillips, C, Elkins, J, Bunkoczi, G, Cooper, C, Pike, A.C.W, Salah, E, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D.A. | | 登録日 | 2007-08-16 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the 3Rd Pdz Domain of Intestine- and Kidney-Enriched Pdz Domain Ikepp (Pdzd3)
To be Published
|
|
1ZNT
 
 | | 18 NMR structures of AcAMP2-Like Peptide with non Natural Fluoroaromatic Residue (AcAMP2F18Pff/Y20Pff) complex with N,N,N-triacetylchitotriose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | 著者 | Chavez, M.I, Andreu, C, Vidal, P, Aboitiz, N, Freire, F, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | 登録日 | 2005-05-12 | | 公開日 | 2005-12-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
2VJ9
 
 | | Human BACE-1 in complex with N-((1S,2R)-3-(cyclohexylamino)-2-hydroxy- 1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl) benzamide | | 分子名称: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-(cyclohexylamino)-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | 著者 | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | 登録日 | 2007-12-07 | | 公開日 | 2008-01-29 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8QEO
 
 | |
8QEN
 
 | |
2YEN
 
 | | Solution structure of the skeletal muscle and neuronal voltage gated sodium channel antagonist mu-conotoxin CnIIIC | | 分子名称: | Mu-conotoxin CnIIIC | | 著者 | Favreau, P, Benoit, E, Hocking, H.G, Carlier, L, D'hoedt, D, Leipold, E, Markgraf, R, Schlumberger, S, Cordova, M.A, Gaertner, H, Paolini-Bertrand, M, Hartley, O, Tytgat, J, Heinemann, S.H, Bertrand, D, Boelens, R, Stocklin, R, Molgo, J. | | 登録日 | 2011-03-28 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel Mu-Conopeptide, Cniiic, Exerts Potent and Preferential Inhibition of Na(V) 1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors.
Br.J.Pharmacol., 166, 2012
|
|
1C57
 
 | | DIRECT DETERMINATION OF THE POSITIONS OF DEUTERIUM ATOMS OF BOUND WATER IN CONCANAVALIN A BY NEUTRON LAUE CRYSTALLOGRAPHY | | 分子名称: | CALCIUM ION, Concanavalin-Br, MANGANESE (II) ION | | 著者 | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb, A.J, Helliwell, J.R. | | 登録日 | 1999-10-26 | | 公開日 | 2000-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | NEUTRON DIFFRACTION (2.4 Å) | | 主引用文献 | Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2WZM
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | 分子名称: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | 著者 | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | 登録日 | 2009-11-30 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2YHT
 
 | |
2V4U
 
 | | Human CTP synthetase 2 - glutaminase domain in complex with 5-OXO-L- NORLEUCINE | | 分子名称: | CTP SYNTHASE 2 | | 著者 | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Weigelt, J, Wikstrom, M, Nordlund, P. | | 登録日 | 2008-09-29 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Human Ctp Synthetase 2 - Glutaminase Domain in Complex with 5-Oxo-L-Norleucine
To be Published
|
|
3B3F
 
 | | The 2.2 A crystal structure of the catalytic domain of coactivator-associated arginine methyl transferase I(CARM1,142-478), in complex with S-adenosyl homocysteine | | 分子名称: | Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
3B72
 
 | | Crystal structure of lysozyme folded in SDS and 2-methyl-2,4-pentanediol | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Michaux, C, Pouyez, J, Wouters, J, Prive, G.G. | | 登録日 | 2007-10-30 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Protecting role of cosolvents in protein denaturation by SDS: a structural study.
Bmc Struct.Biol., 8, 2008
|
|
