4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | 著者 | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | 登録日 | 2015-03-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.916 Å) | | 主引用文献 | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
6WF3
 
 | | Crystal structure of human Naa50 in complex with a cofactor derived inhibitor (compound 1) | | 分子名称: | ACE-MET-LEU-GLY-PRO-NH2, COENZYME A, N-alpha-acetyltransferase 50 | | 著者 | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | 登録日 | 2020-04-03 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFN
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | 分子名称: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | 著者 | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | 登録日 | 2020-04-03 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
4YPE
 
 | | ASH1L SET domain H2193F mutant in complex with S-adenosyl methionine (SAM) | | 分子名称: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | 著者 | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | 登録日 | 2015-03-12 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
4TNI
 
 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | 分子名称: | Putative periplasmic binding protein with substrate ribose | | 著者 | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2015-05-30 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.971 Å) | | 主引用文献 | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
8P8K
 
 | | Acyl-ACP thioesterase from Lemna paucicostata in complex with a thiazolopyridine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-bis(fluoranyl)phenyl]-6-chloranyl-[1,3]thiazolo[4,5-b]pyridine, Acyl-ACP thioesterase | | 著者 | Freigang, J. | | 登録日 | 2023-06-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Study in Scaffold Hopping: Discovery and Optimization of Thiazolopyridines as Potent Herbicides That Inhibit Acyl-ACP Thioesterase.
J.Agric.Food Chem., 71, 2023
|
|
8AQ8
 
 | | FAD-dependent monooxygenase from Stenotrophomonas maltophilia | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Maly, M, Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J. | | 登録日 | 2022-08-11 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Tetracycline-modifying enzyme SmTetX from Stenotrophomonas maltophilia.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8V02
 
 | | AaegOR10 structure bound to o-cresol | | 分子名称: | Odorant receptor OR10, Odorant receptor Orco, o-cresol | | 著者 | Zhao, J, del Marmol, J. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V3D
 
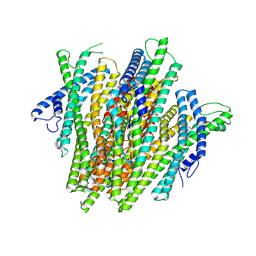 | | AgamOR28 structure bound to 2,4,5-trimethylthiazole | | 分子名称: | 2,4,5-trimethyl-1,3-thiazole, OR28, Odorant receptor Orco | | 著者 | Zhao, J, del Marmol, J. | | 登録日 | 2023-11-27 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V00
 
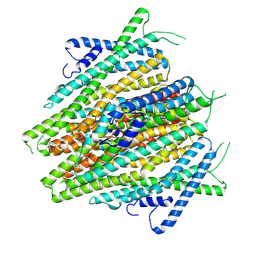 | | AaegOR10 apo structure | | 分子名称: | Odorant receptor OR10, Odorant receptor Orco | | 著者 | Zhao, J, del Marmol, J. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
5JRC
 
 | |
4YPU
 
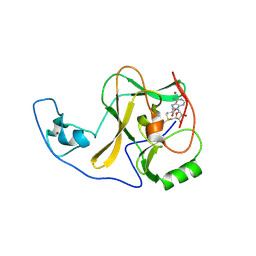 | | ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) | | 分子名称: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | 著者 | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | 登録日 | 2015-03-13 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
8V3C
 
 | |
5IW5
 
 | |
5IZA
 
 | | Protein-protein interaction | | 分子名称: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4TNK
 
 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (5.2 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6WOO
 
 | | CryoEM structure of yeast 80S ribosome with Met-tRNAiMet, eIF5B, and GDP | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal rRNA, ... | | 著者 | Wang, J, Wang, J, Puglisi, J, Fernandez, I.S. | | 登録日 | 2020-04-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | An active role of the eukaryotic large ribosomal subunit in translation initiation fidelity.
To Be Published
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | 分子名称: | NEQ131 | | 著者 | Zhang, J, Gan, J. | | 登録日 | 2016-05-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.900007 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | 分子名称: | CALCIUM ION, Thermolysin, ZINC ION | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6TQR
 
 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | 分子名称: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | 登録日 | 2019-12-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2021-04-11 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
6TRU
 
 | |
