2Y0D
 
 | | BceC mutation Y10K | | Descriptor: | SULFATE ION, UDP-GLUCOSE DEHYDROGENASE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
3TTS
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
1DGI
 
 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2XKA
 
 | |
2XZE
 
 | |
3TQO
 
 | | Structure of the cysteinyl-tRNA synthetase (cysS) from Coxiella burnetii. | | Descriptor: | Cysteinyl-tRNA synthetase, ZINC ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR8
 
 | | Structure of an oligoribonuclease (orn) from Coxiella burnetii | | Descriptor: | ACETATE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2Y8B
 
 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
1DWT
 
 | | Photorelaxed horse heart MYOGLOBIN CO complex | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-12 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
3PTB
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BENZAMIDINE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Bode, W, Schwager, P, Walter, J. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
3PTN
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, TRYPSIN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3QB0
 
 | | Crystal structure of Actin-related protein Arp4 from S. cerevisiae complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 4, CALCIUM ION | | Authors: | Fenn, S, Breitsprecher, D, Gerhold, C.B, Witte, G, Faix, J, Hopfner, K.P. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin.
Embo J., 30, 2011
|
|
3QBY
 
 | | Crystal structure of the PWWP domain of human Hepatoma-derived growth factor 2 | | Descriptor: | H4K20me3 Histone H4 Peptide, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Tempel, W, Amaya, M.F, Adams-Cioaba, M.A, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
2G2K
 
 | | NMR structure of an N-terminal fragment of the eukaryotic initiation factor 5 (eIF5) | | Descriptor: | Eukaryotic translation initiation factor 5 | | Authors: | Conte, M.R, Kelly, G, Babon, J, Sanfelice, D, Smerdon, S.J, Proud, C.G. | | Deposit date: | 2006-02-16 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors
Biochemistry, 45, 2006
|
|
3QKI
 
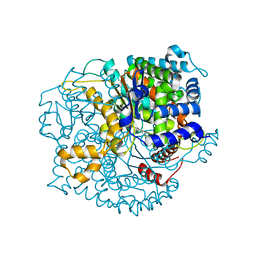 | | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7 | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Gileadi, T, Wernimont, A.K, Hutchinson, A, Weadge, J, Cossar, D, Lew, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7
TO BE PUBLISHED
|
|
2K3K
 
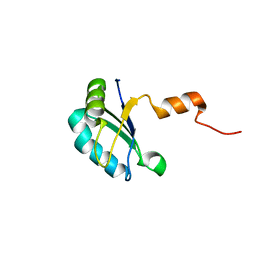 | |
2JDN
 
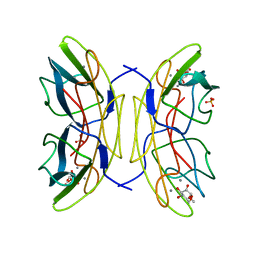 | | Mutant (S22A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-mannopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDU
 
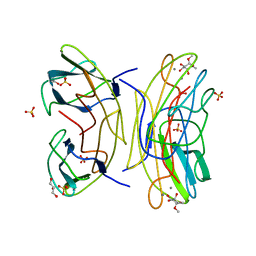 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDY
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-b-D-mannoyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-D-mannopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
1QPL
 
 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-707,587 | | Descriptor: | C32-O-(1-METHYL-INDOL-5-YL) 18-HYDROXY-ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN) | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-25 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
2JDP
 
 | | Mutant (S23A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
