8W83
 
 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8GV0
 
 | | Crystal structure of anti-FIXa IgG fab without FAST-Ig mutations | | Descriptor: | Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
5IX5
 
 | | NMR structure of antibacterial factor-2 | | Descriptor: | Antibacterial factor-related peptide 2 | | Authors: | Masakatsu, K, Umetsu, Y, Rumi, F, Kikukawa, T, Ohki, S, Mizuguchi, M, Demura, M, Aizawa, T. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ABF-2 (antibacterial factor-2) from C. elegans and the interaction with membrane mimetic systems
To Be Published
|
|
4WIB
 
 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
5GLH
 
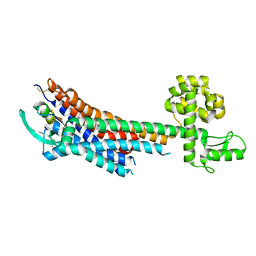 | | Human endothelin receptor type-B in complex with ET-1 | | Descriptor: | Endothelin Receptor Subtype-B, Peptide from Endothelin-1 | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
4X5M
 
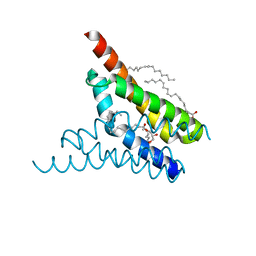 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
7BTW
 
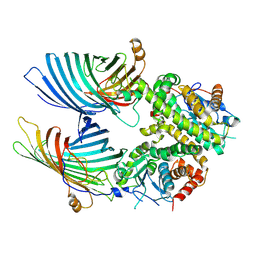 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
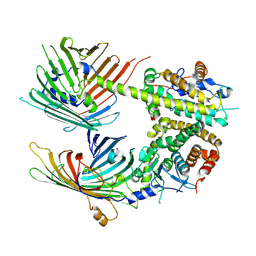 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
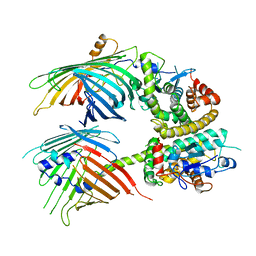 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
8GUJ
 
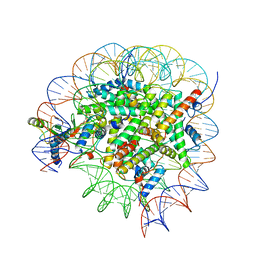 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUK
 
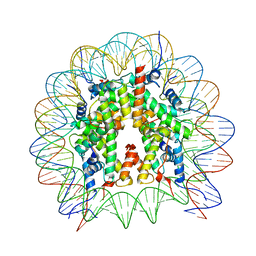 | | Human nucleosome core particle (free form) | | Descriptor: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUI
 
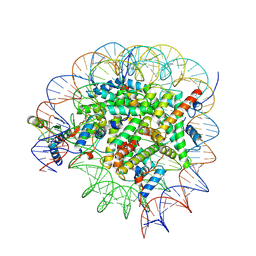 | | Bre1-nucleosome complex (Model I) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8JJS
 
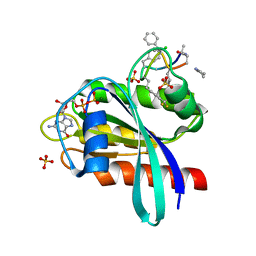 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
3X3C
 
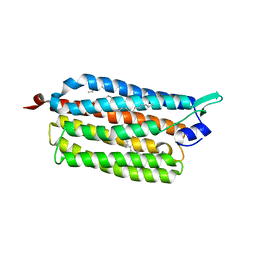 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
3X3B
 
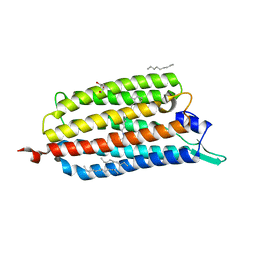 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
6LGP
 
 | | cryo-EM structure of TRPV3 in lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, DIUNDECYL PHOSPHATIDYL CHOLINE, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Shimada, H, Kusakizako, T, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2N92
 
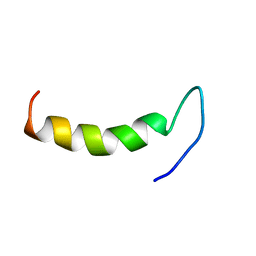 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
2RRF
 
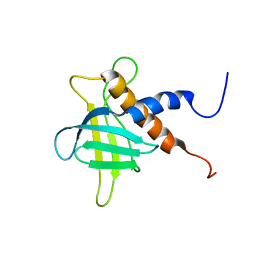 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
8WJ2
 
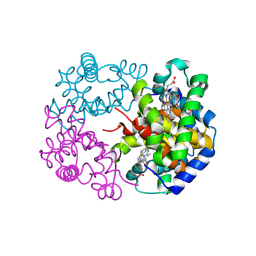 | | cryo-EM structure of human haemoglobin in deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ1
 
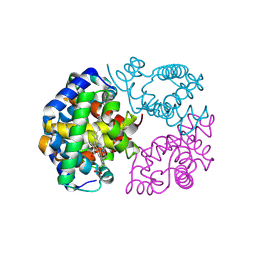 | | cryo-EM structure of human haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIY
 
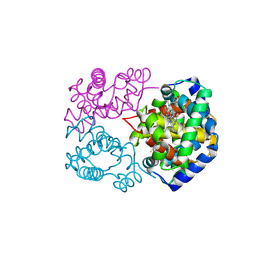 | | cryo-EM structure of alligator haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIZ
 
 | | cryo-EM structure of alligator haemoglobin in deoxy form | | Descriptor: | BICARBONATE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ0
 
 | | cryo-EM structure of human haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
2MDB
 
 | | Tachyplesin I in the presence of lipopolysaccharide | | Descriptor: | Tachyplesin-1 | | Authors: | Kushibiki, T, Kamiya, M, Aizawa, T, Kumaki, Y, Kikukawa, T, Mizuguchi, M, Demura, M, Kawabata, S.I, Kawano, K. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide.
Biochim.Biophys.Acta, 1844, 2014
|
|
