3ATR
 
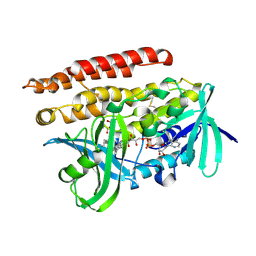 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius co-crystallized with its ligand | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, PYROPHOSPHATE, ... | | Authors: | Fujihashi, M, Sasaki, D, Iwata, Y, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
8JYJ
 
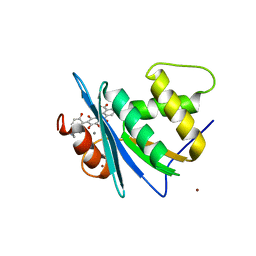 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid A | | Descriptor: | 7-[5-(2-acetamidoethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYH
 
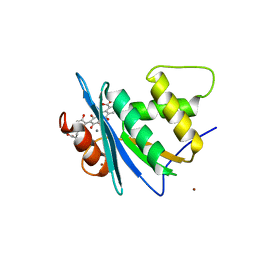 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid C | | Descriptor: | 7-[5-[(2~{S})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYI
 
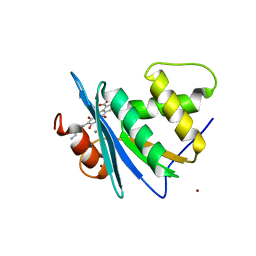 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
1WTH
 
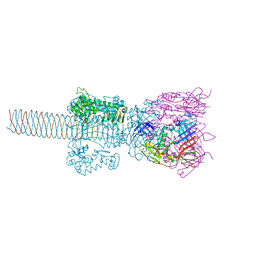 | | Crystal structure of gp5-S351L mutant and gp27 complex | | Descriptor: | Baseplate structural protein Gp27, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kanamaru, S, Ishiwata, Y, Suzuki, T, Rossmann, M.G, Arisaka, F. | | Deposit date: | 2004-11-23 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of bacteriophage t4 tail lysozyme activity during the infection process
J.Mol.Biol., 346, 2005
|
|
5B18
 
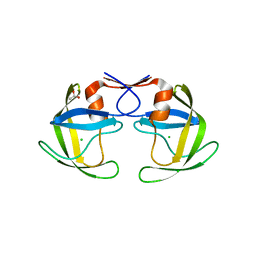 | | Crystal Structure of a Darunavir Resistant HIV-1 Protease | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease | | Authors: | Suzuki, K, Ode, H, Nakashima, M, Sugiura, W, Watanabe, N, Suzuki, A, Iwatani, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique Flap Conformation in an HIV-1 Protease with High-Level Darunavir Resistance
Front Microbiol, 7, 2016
|
|
5Z98
 
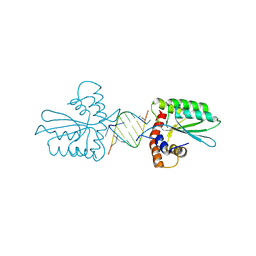 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
3VM8
 
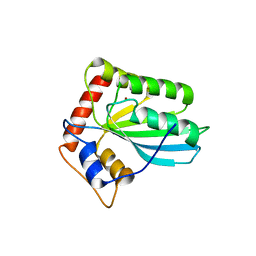 | |
3WUS
 
 | | Crystal Structure of the Vif-Binding Domain of Human APOBEC3F | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Nakashima, M, Kawamura, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Insights into HIV-1 Vif-APOBEC3F Interaction.
J.Virol., 90, 2015
|
|
3VOW
 
 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
