5HZV
 
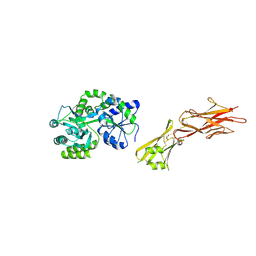 | | Crystal structure of the zona pellucida module of human endoglin/CD105 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Endoglin, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5WR0
 
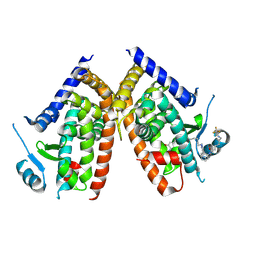 | | Huisgen cycloaddition for PPARg-LBD labeling by soaking method | | Descriptor: | (E)-N-[(3E)-2-oxo-16-(8-{6-[(trifluoroacetyl)amino]hexanoyl}-8,9-dihydro-1H-dibenzo[b,f][1,2,3]triazolo[4,5-d]azocin-1-yl)hexadec-3-en-1-ylidene]glycine, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
2MLQ
 
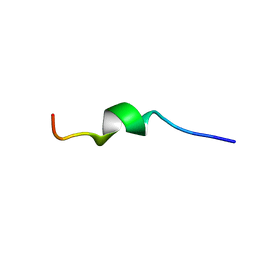 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLO
 
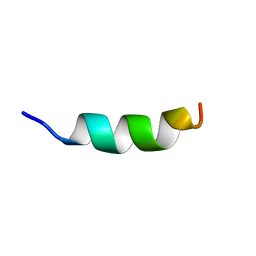 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
5WR1
 
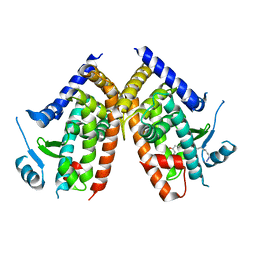 | | Covalent bond formation of bifunctional ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-16-azido-2-oxidanylidene-hexadec-3-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
5WQX
 
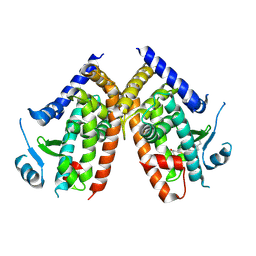 | | Covalent bond formation of synthetic ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-2-oxidanylidenehexadec-5-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
5XPO
 
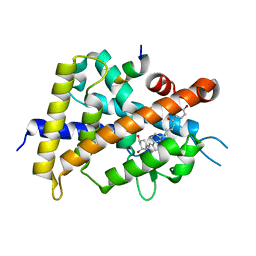 | | Crystal structure of VDR-LBD complexed with 25-(hydroxyphenyl)-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (5~{R})-5-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexyl idene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-(4-hydroxyphenyl)hexan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
7CMQ
 
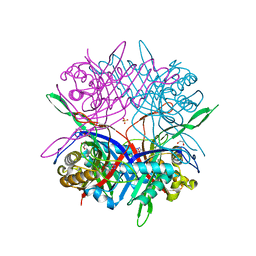 | |
2D81
 
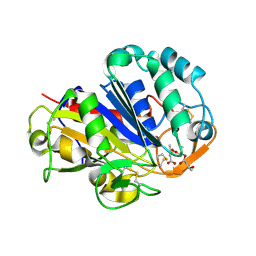 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D80
 
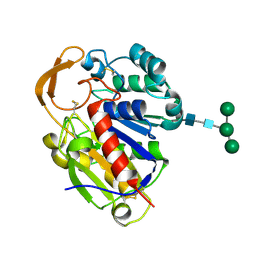 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
7CMN
 
 | |
7CUF
 
 | |
8HHQ
 
 | |
7CUG
 
 | |
8HHP
 
 | |
7CUC
 
 | |
5H1E
 
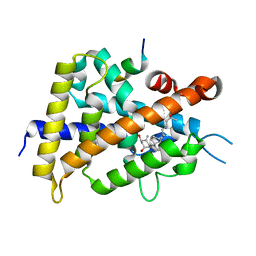 | | Interaction between vitamin D receptor and coactivator peptide SRC2-3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 2 peptide, Vitamin D3 receptor | | Authors: | Egawa, D, Itoh, T, Kato, A, Kataoka, S, Anami, Y, Yamamoto, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SRC2-3 binds to vitamin D receptor with high sensitivity and strong affinity
Bioorg. Med. Chem., 25, 2017
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
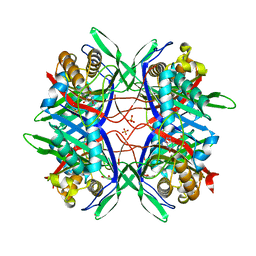
5YJA
 
 | |
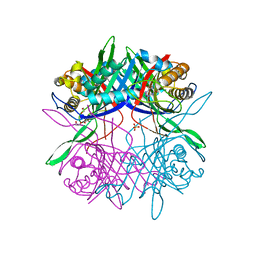
5Z2B
 
 | |
3E9H
 
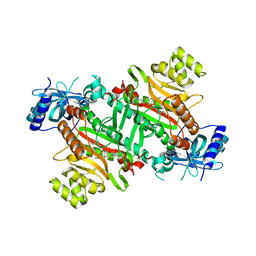 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysylsulfamoyl adenosine | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
5Z27
 
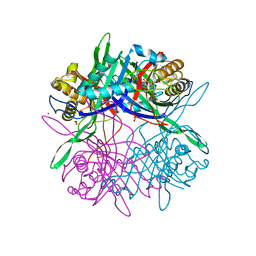 | |
2MWI
 
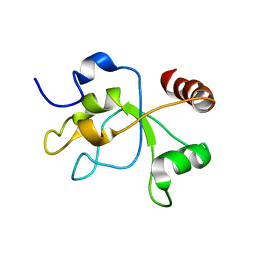 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
3E9I
 
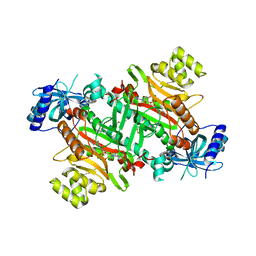 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysine hydroxamate-AMP | | Descriptor: | 5'-O-{(R)-hydroxy[(L-lysylamino)oxy]phosphoryl}adenosine, Lysyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
5ZJG
 
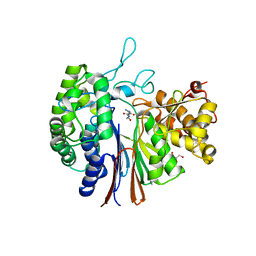 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with Gly-Gly | | Descriptor: | GLYCEROL, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 L-subunit, ... | | Authors: | Hibi, T, Imaoka, M, Itoh, T, Wakayama, M. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure analysis and enzymatic characterization of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
