5YJA
 
 | |
5Z2B
 
 | |
5HZV
 
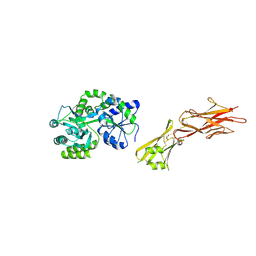 | | Crystal structure of the zona pellucida module of human endoglin/CD105 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Endoglin, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
5AZT
 
 | | Ternary complex of hPPARalpha ligand binding domain, 17-oxoDHA and a SRC1 peptide | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
5Z27
 
 | |
5AWK
 
 | | Crystal structure of VDR-LBD/partial agonist complex: 22S-ethyl analogue | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-ethyl-5-oxidanyl-pentan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor,Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-07-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Bioorg.Med.Chem., 23, 2015
|
|
5AZV
 
 | | Crystal structure of hPPARgamma ligand binding domain complexed with 17-oxoDHA | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
5B41
 
 | | Crystal structure of VDR-LBD complexed with 2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Apo- and Antagonist-Binding Structures of Vitamin D Receptor Ligand-Binding Domain Revealed by Hybrid Approach Combining Small-Angle X-ray Scattering and Molecular Dynamics
J.Med.Chem., 59, 2016
|
|
3E9H
 
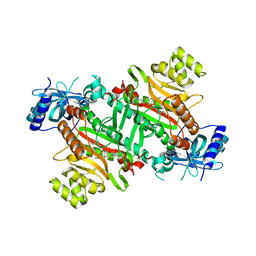 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysylsulfamoyl adenosine | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
5ZWI
 
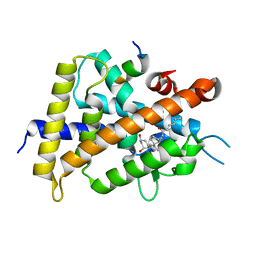 | | Interaction between Vitamin D receptor (VDR) and a ligand having a dienone group | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWH
 
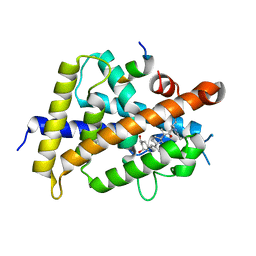 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZJG
 
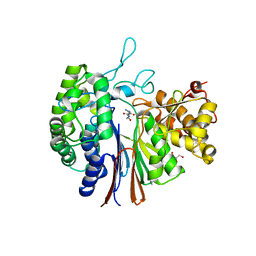 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with Gly-Gly | | Descriptor: | GLYCEROL, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 L-subunit, ... | | Authors: | Hibi, T, Imaoka, M, Itoh, T, Wakayama, M. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure analysis and enzymatic characterization of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
5ZWE
 
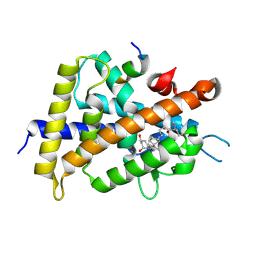 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
6A6U
 
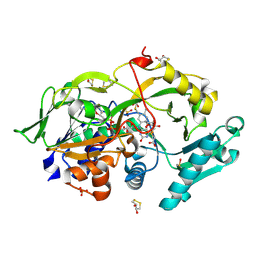 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation, in complex with FSA | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6T
 
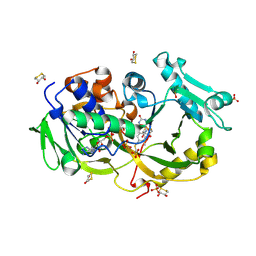 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6V
 
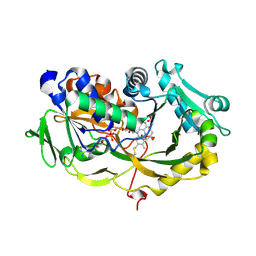 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with 7 additional mutations, in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
5B5B
 
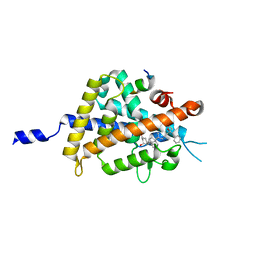 | | Crystal structure of VDR-LBD complexed with 2-methylidene-26,27-diphenyl-19-nor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-oxidanyl-7-phenyl-6-(phenylmethyl)heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix12-Stabilization Antagonist of Vitamin D Receptor
Bioconjug.Chem., 27, 2016
|
|
6A6R
 
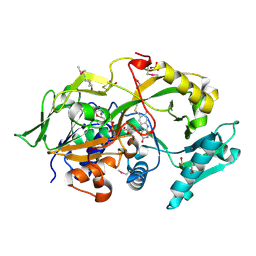 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6S
 
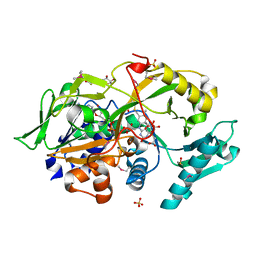 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans in complex with FSA, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
5ZWF
 
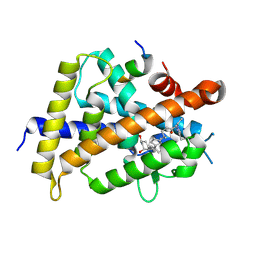 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a enone with a beta methyl group via conjugate addition reaction | | Descriptor: | (E,7R)-7-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-2-en-4-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5XUQ
 
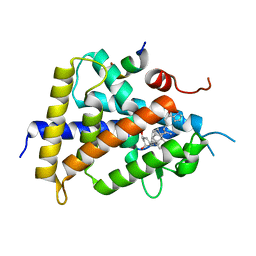 | | Crystal structure of VDR-LBD complexed with an antagonist, 2-methylidene-19,26,27-trinor-22-(S)-butyl-1-hydroxy-25-oxo-25-(1H-pyrrol-2-yl)- vitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(1~{H}-pyrrol-2-yl)octan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent Vitamin D Receptor Antagonist
To Be Published
|
|
2H84
 
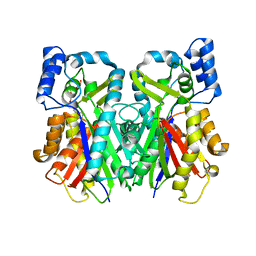 | | Crystal Structure of the C-terminal Type III Polyketide Synthase (PKS III) Domain of 'Steely1' (a Type I/III PKS Hybrid from Dictyostelium) | | Descriptor: | HEXAETHYLENE GLYCOL, Steely1 | | Authors: | Austin, M.B, Saito, T, Bowman, M.E, Haydock, S, Kato, A, Moore, B.S, Kay, R.R, Noel, J.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase.
Nat.Chem.Biol., 2, 2006
|
|
5H1E
 
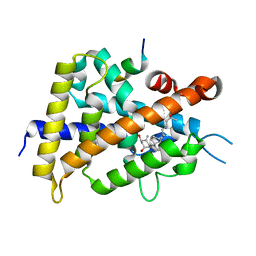 | | Interaction between vitamin D receptor and coactivator peptide SRC2-3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 2 peptide, Vitamin D3 receptor | | Authors: | Egawa, D, Itoh, T, Kato, A, Kataoka, S, Anami, Y, Yamamoto, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SRC2-3 binds to vitamin D receptor with high sensitivity and strong affinity
Bioorg. Med. Chem., 25, 2017
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
