4YVG
 
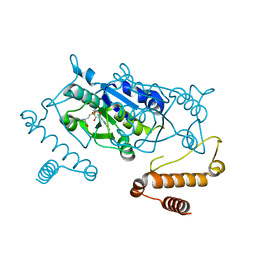 | | Crystal Structure of H. influenzae TrmD in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Ito, T, Yokoyama, S. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
9KS7
 
 | |
9KPL
 
 | | Crystal structure of T. rubripes Mincle with glucose | | Descriptor: | C-type lectin domain-containing protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Ito, T, Nagae, M, Yamasaki, S. | | Deposit date: | 2024-11-23 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetic and structural insights into the origin of C-type lectin Mincle in vertebrates.
Immunogenetics, 77, 2025
|
|
4H04
 
 | | Lacto-N-biosidase from Bifidobacterium bifidum | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ito, T, Katayama, T, Wada, J, Suzuki, R, Ashida, H, Wakagi, T, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2012-09-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
5BXP
 
 | | LNBase in complex with LNB-LOGNAc | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXS
 
 | | LNBase in complex with LNB-NHAcCAS | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
4JAW
 
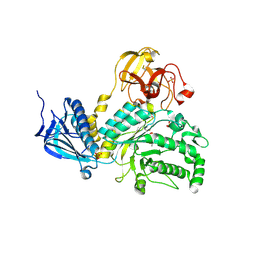 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
1Q8K
 
 | |
1U2F
 
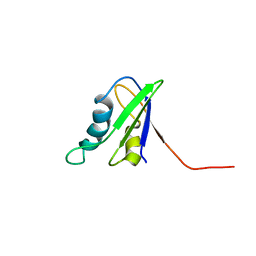 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
3AFH
 
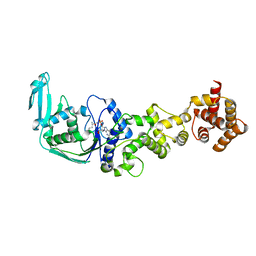 | |
3AKZ
 
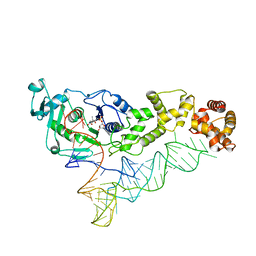 | |
3AL0
 
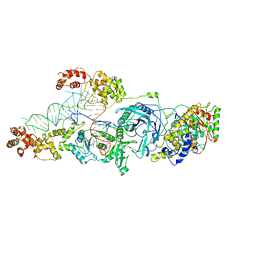 | | Crystal structure of the glutamine transamidosome from Thermotoga maritima in the glutamylation state. | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C,Linker,Glutamate--tRNA ligase 2, ... | | Authors: | Ito, T, Yokoyama, S. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.368 Å) | | Cite: | Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
Nature, 467, 2010
|
|
3WKW
 
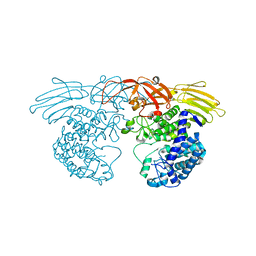 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum ligand free form | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3WKX
 
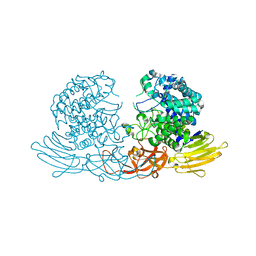 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum arabinose complex form | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
2U2F
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
5GZT
 
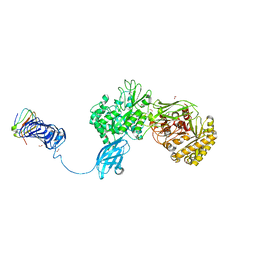 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5GZU
 
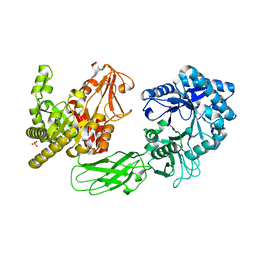 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5I05
 
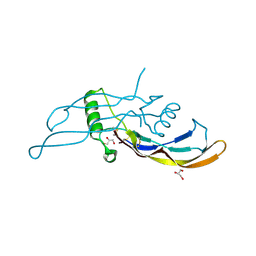 | |
1FP3
 
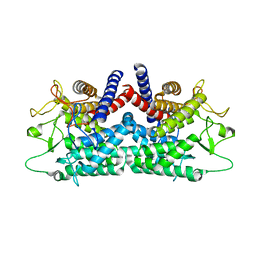 | | CRYSTAL STRUCTURE OF N-ACYL-D-GLUCOSAMINE 2-EPIMERASE FROM PORCINE KIDNEY | | Descriptor: | N-ACYL-D-GLUCOSAMINE 2-EPIMERASE | | Authors: | Itoh, T, Mikami, B, Maru, I, Ohta, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2000-08-30 | | Release date: | 2000-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
J.Mol.Biol., 303, 2000
|
|
5GZV
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5I04
 
 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7C7D
 
 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8W7P
 
 | | Extracellular domain of a sensor histidine kinase | | Descriptor: | Extracellular domain of a sensor histidine kinase NagS, GLYCEROL | | Authors: | Itoh, T, Ogawa, T, Hibi, T, Kimoto, H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of the extracellular domain of sensor histidine kinase NagS from Paenibacillus sp. str. FPU-7: nagS interacts with oligosaccharide binding protein NagB1 in complexes with N, N'-diacetylchitobiose.
Biosci.Biotechnol.Biochem., 88, 2024
|
|
