2CZI
 
 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with calcium and phosphate ions | | Descriptor: | CALCIUM ION, Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Arai, R, Ito, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZE
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Arai, R, Ito, K, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP
To be Published
|
|
2CZD
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
2CZ5
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3
To be Published
|
|
5B1A
 
 | | Bovine heart cytochrome c oxidase in the fully oxidized state at 1.5 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
5B1B
 
 | | Bovine heart cytochrome c oxidase in the fully reduced state at 1.6 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
7COY
 
 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
6LX8
 
 | | X-ray structure of human PPARalpha ligand binding domain-oleic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, OLEIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX7
 
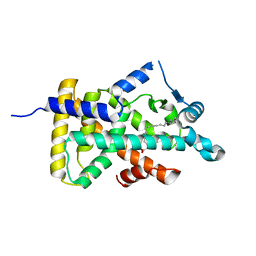 | | X-ray structure of human PPARalpha ligand binding domain-stearic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, STEARIC ACID | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5JRB
 
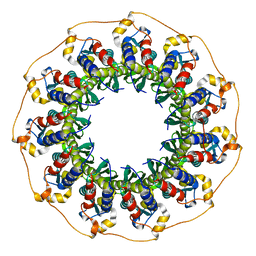 | | Rad52(1-212) K102A/K133A/E202A mutant | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Saotome, M, Saito, K, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2016-05-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the human DNA-repair protein RAD52 containing surface mutations.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7VKF
 
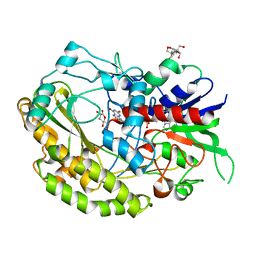 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VKD
 
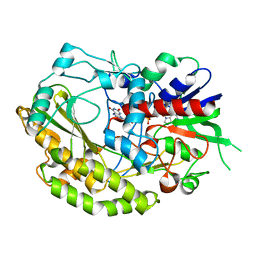 | |
1G3D
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE COPPER (II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3B
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE MAGNESIUM(II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3C
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF BASE IRON(III) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3E
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF-BASE COPPER (II) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
5IWA
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with the GE81112 peptide antibiotic | | Descriptor: | (2S,3S)-2-{[(2S)-3-(2-amino-1H-imidazol-5-yl)-2-{[(2S,4S)-5-(carbamoyloxy)-4-hydroxy-2-({[(2S,3S)-3-hydroxypiperidin-2-yl]carbonyl}amino)pentanoyl]amino}propanoyl]amino}-3-(2-chloro-1H-imidazol-5-yl)-3-hydroxypropanoic acid, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Chieko, N, Masahito, K, Takemoto, C, Yokoyama, S, Connell, S.R, Fucini, P. | | Deposit date: | 2016-03-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BRO
 
 | | Crystal structure of modified HexB (modB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | Authors: | Kitakaze, K, Maita, N, Itoh, K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
8JX2
 
 | | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer | | Descriptor: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | Authors: | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
8JX3
 
 | | alpha-Hemolysin(G122S/K147R/K237C)-SpyTag/SpyCatcher head to head 14-mer | | Descriptor: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | Authors: | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
7COH
 
 | | Dimeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2020-08-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3-A Resolution structure of bovine cytochrome c oxidase suggests a dimerization mechanism
Biochim.Biophys.Acta, 2021
|
|
1V55
 
 | | Bovine heart cytochrome c oxidase at the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V54
 
 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
4QSE
 
 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum c58, target efi-510558, with bound glycerol | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, K.L, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of maltoside transporter ATU4361 from
Agrobacterium Fabrum, target EFI-510558
To be Published
|
|
8H8R
 
 | | Bovine Heart Cytochrome c Oxidase in the Calcium-bound Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calcium-bound structure of bovine cytochrome c oxidase.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
