7MMN
 
 | |
6H2Y
 
 | | human Fab 1E6 bound to fHbp variant 3 from Neisseria meningitidis serogroup B | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Veggi, D, Bianchi, F, Cozzi, R, Malito, E, Bottomley, M.J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.
Faseb J., 33, 2019
|
|
7MPG
 
 | |
7SBZ
 
 | |
7SA6
 
 | | fHbp mutant 2416 bound to Fab JAR5 | | Descriptor: | Factor H-binding protein 2416, JAR5 Heavy Chain, JAR5 Light Chain | | Authors: | Chesterman, C, Malito, E, Bottomley, M.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active Learning for Rapid Design: An iterative AI approach for accelerated vaccine design that combines active machine learning and high-throughput experimental evaluation
To Be Published
|
|
7NRU
 
 | |
4NBY
 
 | | Crystal Structure of TcdA-A2 Bound to Two Molecules of A20.1 VHH | | Descriptor: | A20.1 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC1
 
 | | Crystal Structure of TcdA-A2 Bound to A20.1 VHH and A26.8 VHH | | Descriptor: | A20.1 VHH, A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
3CQF
 
 | | Crystal structure of anthrolysin O (ALO) | | Descriptor: | Thiol-activated cytolysin | | Authors: | Bourdeau, R.W, Malito, E, Tang, W.J. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cellular Functions and X-ray Structure of Anthrolysin O, a Cholesterol-dependent Cytolysin Secreted by Bacillus anthracis
J.Biol.Chem., 284, 2009
|
|
2X9W
 
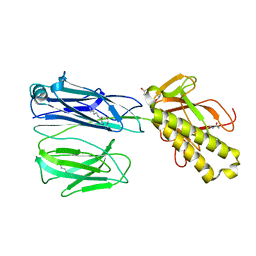 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
4OXR
 
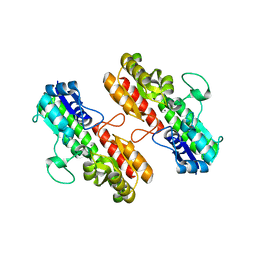 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | Descriptor: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
4OXQ
 
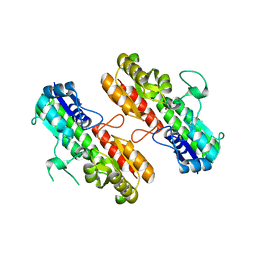 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | Descriptor: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
2X9Z
 
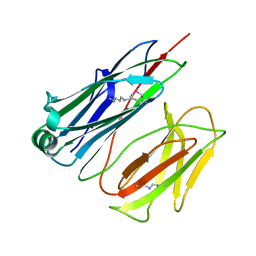 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Y
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9X
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
4NBZ
 
 | | Crystal Structure of TcdA-A1 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
2VQB
 
 | | Bacterial flavin-containing monooxygenase in complex with NADP: soaking in aerated solution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alfieri, A, Malito, E, Orru, R, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Revealing the Moonlighting Role of Nadp in the Structure of a Flavin-Containing Monooxygenase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WC0
 
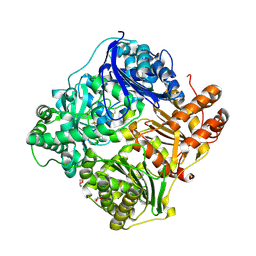 | | crystal structure of human insulin degrading enzyme in complex with iodinated insulin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Manolopoulou, M, Guo, Q, Malito, E, Schilling, A.B, Tang, W.J. | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Basis of Catalytic Chamber-Assisted Unfolding and Cleavage of Human Insulin by Human Insulin Degrading Enzyme.
J.Biol.Chem., 284, 2009
|
|
2VQ7
 
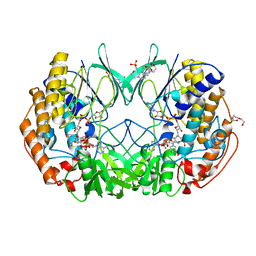 | | Bacterial flavin-containing monooxygenase in complex with NADP: native data | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alfieri, A, Malito, E, Orru, R, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revealing the Moonlighting Role of Nadp in the Structure of a Flavin-Containing Monooxygenase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WBY
 
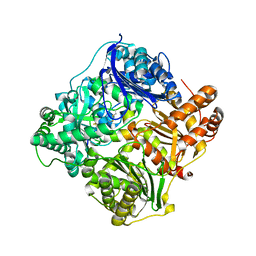 | | Crystal structure of human insulin-degrading enzyme in complex with insulin | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, INSULIN-DEGRADING ENZYME, ... | | Authors: | Manolopoulou, M, Guo, Q, Malito, E, Schilling, A.B, Tang, W.J. | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of Catalytic Chamber-Assisted Unfolding and Cleavage of Human Insulin by Human Insulin Degrading Enzyme.
J.Biol.Chem., 284, 2009
|
|
4NC0
 
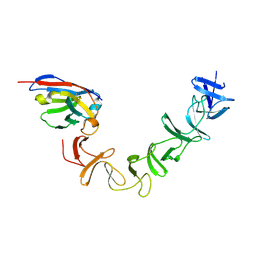 | | Crystal Structure of TcdA-A2 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC2
 
 | | Crystal structure of TcdB-B1 bound to B39 VHH | | Descriptor: | B39 VHH, Toxin B | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NBX
 
 | | Crystal Structure of Clostridium difficile Toxin A fragment TcdA-A1 Bound to A20.1 VHH | | Descriptor: | A20.1 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
