3KKM
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKP
 
 | | Crystal structure of M-Ras P40D in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
1FIY
 
 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QTR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PROLYL AMINOPEPTIDASE FROM SERRATIA MARCESCENS | | Descriptor: | PROLYL AMINOPEPTIDASE | | Authors: | Yoshimoto, T, Kabashima, T, Uchikawa, K, Inoue, T, Tanaka, N. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of prolyl aminopeptidase from Serratia marcescens.
J.Biochem.(Tokyo), 126, 1999
|
|
8K9N
 
 | | Subatomic resolution structure of Pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Lintuluoto, M, Kurihara, K, Hasegawa, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
8K9P
 
 | | Neutron X-ray joint structure of pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Kurihara, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
1BYO
 
 | | WILD-TYPE PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1BYP
 
 | | E43K,D44K DOUBLE MUTANT PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1MGT
 
 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | Descriptor: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | Authors: | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
8GZ6
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nanobody P17 | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
8GZ5
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
5H5H
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
4YSC
 
 | | Completely oxidized structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H5G
 
 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
4YSE
 
 | | High resolution synchrotron structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H5I
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
1MSF
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
1MSE
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
8H1O
 
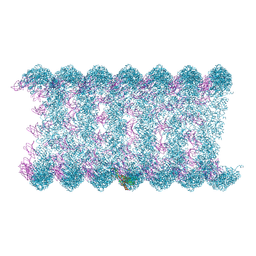 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8IBN
 
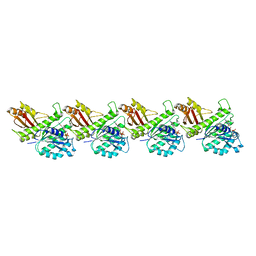 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
1MHO
 
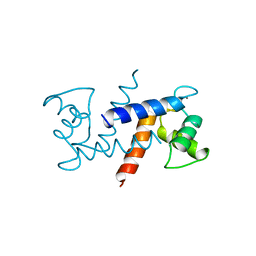 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
