3RG6
 
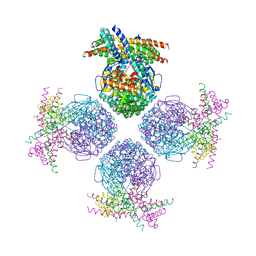 | | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco | | Descriptor: | RbcX protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Bracher, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4IRW
 
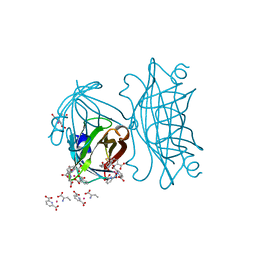 | | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form | | Descriptor: | BIOTIN, PYRIDINE-2,6-DICARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Bandara, R.A.M.S.S, Liu, D.Q, Hindupur, A, Tesh, K.F, Fox, R.O. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form
To be Published
|
|
6GHK
 
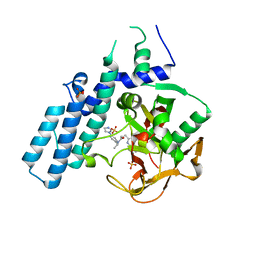 | | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527 | | Descriptor: | Poly [ADP-ribose] polymerase 1, SULFATE ION, ~{N}-[(1~{R})-1-(4-imidazol-1-ylphenyl)ethyl]-3-(4-oxidanylidene-1~{H}-quinazolin-2-yl)propanamide | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Moche, M, Brock, J, Ekblad, T, Spjut, S, Elofsson, M, Schuler, H. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527
To Be Published
|
|
6GMH
 
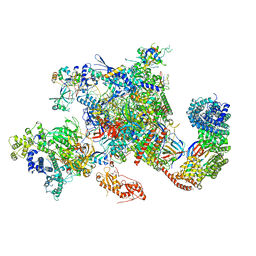 | | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6 | | Descriptor: | CDC73, CTR9,RNA polymerase-associated protein CTR9 homolog,RNA polymerase-associated protein CTR9 homolog, DNA-directed RNA polymerase II subunit RPB9, ... | | Authors: | Vos, S.M, Farnung, L, Boehing, M, Linden, A, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2018-05-26 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6.
Nature, 560, 2018
|
|
4KB4
 
 | | Crystal structure of ribosome recycling factor mutant R31A from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KDD
 
 | | Structure of Mycobacterium tuberculosis ribosome recycling factor in presence of detergent | | Descriptor: | CADMIUM ION, DECYL-BETA-D-MALTOPYRANOSIDE, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
2CI8
 
 | | sh2 domain of human nck1 adaptor protein - uncomplexed | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
4L7L
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0368 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(2S)-1-hydroxy-3-phenylpropan-2-yl]-3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L7U
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0398 | | Descriptor: | Poly [ADP-ribose] polymerase 3, methyl N-[3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanoyl]-L-phenylalaninate | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L6Z
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor STO1168 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-(pyridin-4-yl)ethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L7O
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor STO1542 | | Descriptor: | DIMETHYL SULFOXIDE, N-{(1S)-1-[4-(1H-imidazol-1-yl)phenyl]ethyl}-3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L7R
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0400 | | Descriptor: | N-[(2S)-1-hydroxybutan-2-yl]-3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L70
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0352 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-phenylpropyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L7P
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0395 | | Descriptor: | (2E)-N-[(2S)-1-hydroxy-3-phenylpropan-2-yl]-3-(4-oxo-1,4-dihydroquinazolin-2-yl)prop-2-enamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4L7N
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor STO1542 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-(4-sulfamoylphenyl)ethyl]propanamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
4KB2
 
 | | Crystal structure of ribosome recycling factor mutant R109A from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KC6
 
 | | Crystal structure of C-terminal deletion mutant of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KAW
 
 | | Crystal structure of ribosome recycling factor mutant R39G from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
2CI9
 
 | | Nck1 SH2-domain in complex with a dodecaphosphopeptide from EPEC protein Tir | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
1KYQ
 
 | | Met8p: A bifunctional NAD-dependent dehydrogenase and ferrochelatase involved in siroheme synthesis. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Siroheme biosynthesis protein MET8 | | Authors: | Schubert, H.L, Raux, E, Brindley, A.A, Wilson, K.S, Hill, C.P, Warren, M.J. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase.
EMBO J., 21, 2002
|
|
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
2WVW
 
 | | Cryo-EM structure of the RbcL-RbcX complex | | Descriptor: | RBCX PROTEIN, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN | | Authors: | Liu, C, Young, A.L, Starling-Windhof, A, Bracher, A, Saschenbrecker, S, Rao, B.V, Rao, K.V, Berninghausen, O, Mielke, T, Hartl, F.U, Beckmann, R, Hayer-Hartl, M. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Coupled Chaperone Action in Folding and Assembly of Hexadecameric Rubisco
Nature, 463, 2010
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
