8UZE
 
 | | Crystal structure of chimeric bat coronavirus BANAL-20-236 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of chimeric RBD complexed with chimeric mouse ACE2
To Be Published
|
|
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7KM6
 
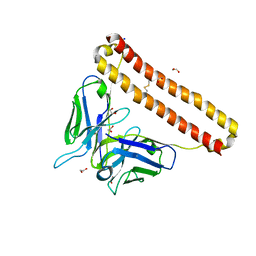 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
5XFA
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
7RWN
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWP
 
 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
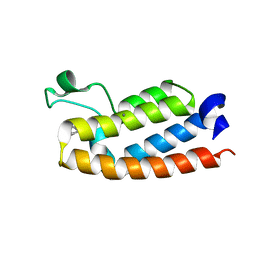 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
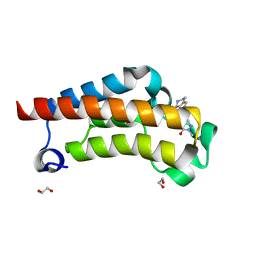 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6U81
 
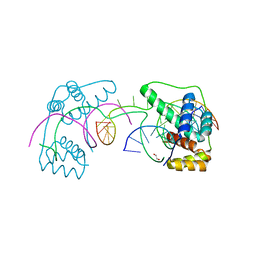 | | Crystal Structure of the Double Homeodomain of DUX4 in Complex with a DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*CP*CP*CP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2019-09-04 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA aptamers against the DUX4 protein reveal novel therapeutic implications for FSHD.
Faseb J., 34, 2020
|
|
8VQR
 
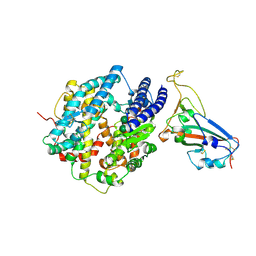 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
5XF9
 
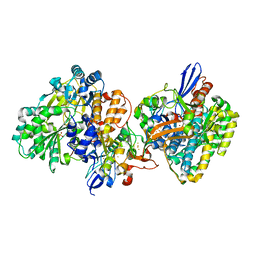 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
7MLR
 
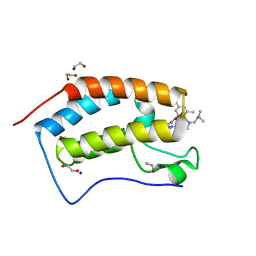 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1- yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (DW34) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
1IQ5
 
 | | Calmodulin/nematode CA2+/Calmodulin dependent kinase kinase fragment | | Descriptor: | CA2+/CALMODULIN DEPENDENT KINASE KINASE, CALCIUM ION, CALMODULIN | | Authors: | Kurokawa, H, Osawa, M, Kurihara, H, Katayama, N, Tokumitsu, H, Swindells, M.B, Kainosho, M, Ikura, M. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide
J.Mol.Biol., 312, 2001
|
|
4NQ3
 
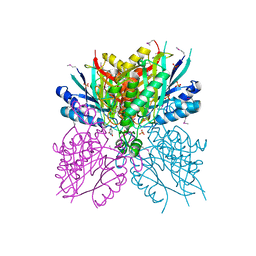 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | Descriptor: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | Authors: | Cho, S, Shi, K, Aihara, H. | | Deposit date: | 2013-11-23 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
1Z1G
 
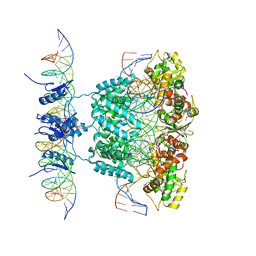 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | Descriptor: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
7K30
 
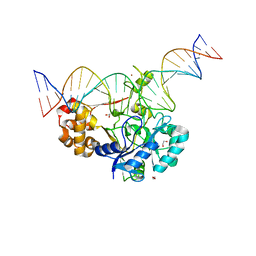 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
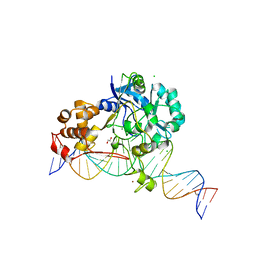 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6B0B
 
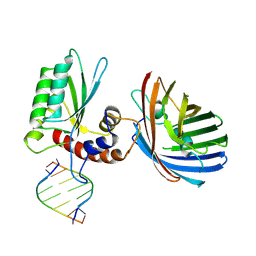 | | Crystal structure of human APOBEC3H | | Descriptor: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.2800622 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
6P7B
 
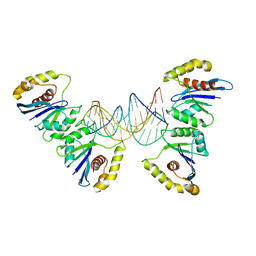 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
8FR5
 
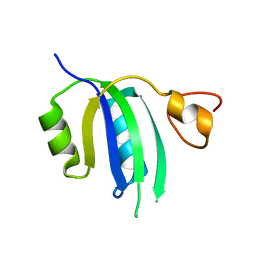 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7TV2
 
 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
5SWW
 
 | | Crystal Structure of Human APOBEC3A complexed with ssDNA | | Descriptor: | DNA 15-Mer, DNA dC->dU-editing enzyme APOBEC-3A, GLYCEROL, ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8E5E
 
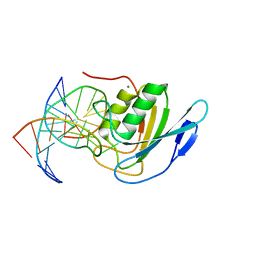 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine flipped into the active site | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, ... | | Authors: | Yin, L.L, Shi, K, Aihara, H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7MC6
 
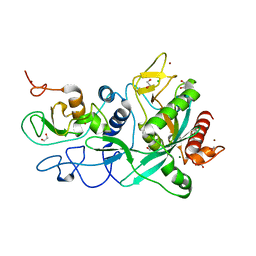 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
